In drug development, it isn’t easy to learn molecular representation from chemical structures since traditional molecular representation techniques, such as sequence-based and graph-based approaches, have limits in extracting useful vectors due to their extensive domain knowledge. Inspired by deep learning technologies based on images and computer vision, researchers have developed a self-supervised framework for learning picture representations that integrates protein and chemical image representations to accurately predict compound-protein interactions. IBM, Cleveland Clinic, and Kent State University have collaborated to create a drug discovery AI algorithm for treating chronic pain.
Introduction
In the US, 50 million adults suffer from chronic pain, and 20% of people worldwide. Opioids, which are popular analgesics, have unfavorable side effects that include drug addiction. The need for less addictive non-opioid analgesics is highlighted by the opioid crisis. Since they cause intracellular signaling events in sensory neurons, G-protein-coupled receptors (GPCRs) are frequently targeted for pain management. Biased agonists of opioids or other GPCRs have been discovered recently to prevent negative effects. It is still difficult to identify unique chemotypes that produce analgesia without causing drug addiction side effects. There has been evidence linking the gut microbiota and its metabolites to the morbidity of chronic pain, with long-term pain being associated with a decrease in the amount of short-chain fatty acids.
The identification of prospective medications and drug repurposing has shown promise with recent developments in artificial intelligence (AI)-based compound-protein interaction (CPI) predictions. Handcrafted molecular fingerprint descriptors and protein sequence descriptors have been trained using a variety of traditional machine-learning algorithms, including support vector machine, random forest, and kernel regression. However, the low dimensionality of these descriptors frequently prevents them from capturing pharmacologically significant information. An unsupervised deep learning system, ImageMol, was developed to enhance the accuracy of CPI predictions by extracting pharmacologically relevant properties of ligands from molecular image representations, surpassing traditional sequence-based and graph-based models. With the help of amino acid sequences, the AlphaFold2 model can predict the structures of the whole human proteome in a systematic manner, indicating the possibility of using three-dimensional (3D) structural data to predict CPI.
Key Features of the Study
- Deep learning and artificial intelligence (AI) technologies have the potential to find effective medications for treating human illnesses, including pain.
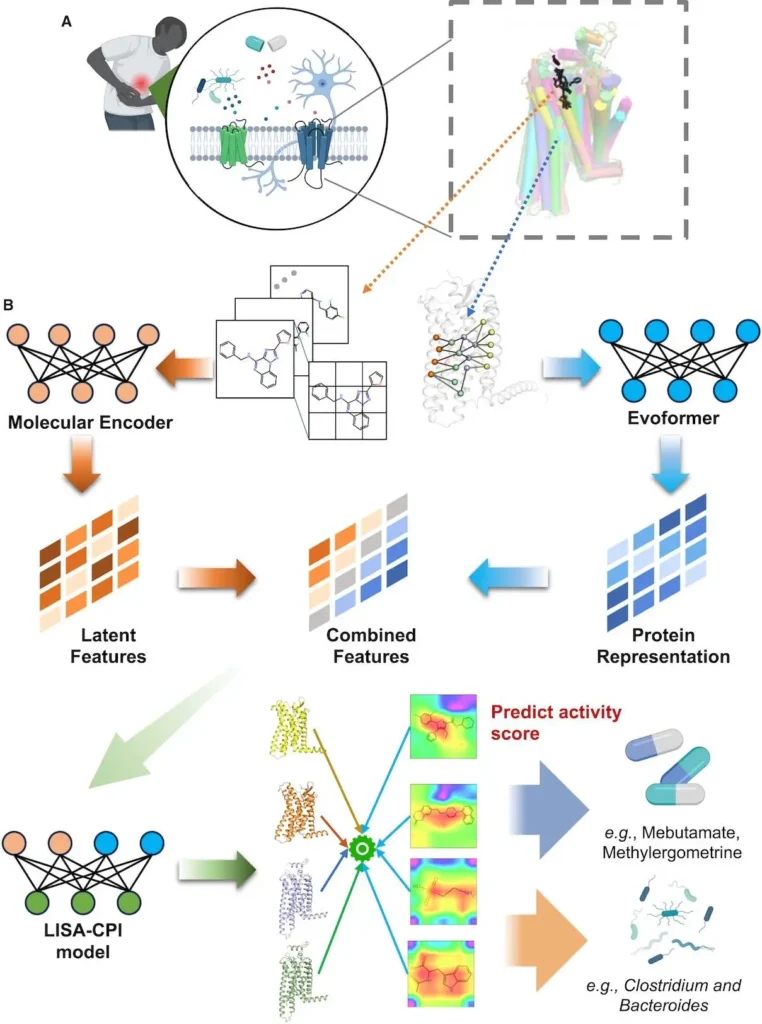
- Here, researchers describe an interpretable deep-learning-based framework to predict compound-protein interactions (LISA-CPI) that takes into account the three-dimensional (3D) structure of receptors and ligand images. An enhanced AlphaFold2-based technique (Evoformer) and an unsupervised deep-learning-based molecular picture representation (ImageMol) of ligands are integrated by LISA-CPI.
- On experimental CPIs connecting 104,969 ligands and 33 G-protein-coupled receptors (GPCRs), researchers showed that LISA-CPI produced an improvement of about 20% in the average mean absolute error (MAE) on par with state-of-the-art models.
- Utilizing LISA-CPI, scientists were able to rank prospective repurposable medications (like methylergometrine) and discover candidate gut-microbiota-derived metabolites (like citicoline) that may be used to treat pain by selectively binding to human GPCRs.
- Researchers have demonstrated that, when combined with a deep learning framework, the integration of molecular imaging and protein 3D structural representations offers a potent computational drug development method for treating pain and other complicated disorders.
Collaboration of Academia and Industry
An AI drug discovery method for treating chronic pain has been developed in partnership with Kent State University and IBM. G protein-coupled receptors (GPCRs), which can relieve pain without becoming addicted or requiring the use of opioids, are the objective of the study, which is being conducted under the direction of postdoctoral associate Yunguang Qiu. Finding FDA-approved medications for prospective pain indications is one of the research approaches already developed by the Cheng Lab, which focuses on producing treatments for nervous system problems. The group has been leading a team to improve an earlier AI system created by the Cheng Lab, and one of its members is computational scientist Yuxin Yang, a former doctoral student at Kent State University. The researchers have been able to collaborate with and learn from colleagues in the industry sector thanks to the invaluable guidance and perspective that IBM collaborators have provided in developing sophisticated computational methodologies.
More About the Work
In this work, scientists introduce a deep learning framework (called LISA-CPI) to predict CPIs by combining representations based on protein 3D structures and ligand images. With chemical awareness and 3D protein residue pair representations, the method surpassed existing methods on CPI prediction of GPCR and kinase benchmarks (ImageMol). Researchers used LISA-CPI to predict possible medications from US Food and Drug Administration (FDA)-)-approved pharmaceuticals and gut-microbiota-derived metabolites to find potential pain treatment strategies. Therefore, by precisely targeting pain-associated GPCRs, they could highlight possible repurposable medicines (like methylergometrine) and gut metabolite-based (like citicoline) candidate treatments for pain. In conclusion, if widely used, the LISA-CPI paradigm provides a helpful computational drug discovery approach for pain and other human disorders.
Conclusion
The LISA-CPI framework is a deep learning-based drug discovery framework that combines protein 3D structure representation for binding activity prediction using ligand-GPCR interactions with molecular image representation for ligands. This framework achieves minimal computational cost and accuracy by utilizing the pre-trained Evoformer from AlphaFold2 and the pre-trained molecular encoder from ImageMol. For every compound-protein pair, the LISA-CPI framework manages both molecular images and protein structure representations, enabling it to predict binding affinity and functioning without requiring knowledge of structural binding site details. The framework predicts binding activities and functional predictions better than other cutting-edge molecular representation models, such as sequence-based and graph-based models.
Article Source: Reference Article | Reference Article
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.