Bioinformatics, in its continuous advancement, depends on the detailed examination of proteins within its molecular domain. Proteins, the working structures of the cell, have a hand in nearly all intracellular activities. Depending on their structures and the relationships between them, all of their functions determine why and how the investigation of proteins is paramount in medicine, biotechnology, and the scientific comprehension of life. Westlake University’s ProTrek, a tri-modal protein language model, SFF transforms protein research by supporting contrastive learning of protein signs, signals, forms, and functions.
Navigating the Protein Universe: A Challenging Landscape
The protein environment is rich and complex, posing significant problems to researchers due to the complicated structure and several roles in metabolic processes. Gaining insights into the three attributes of proteins – primary structure, secondary structure, and tertiary structure and function (SSF) is crucial to understanding life itself. ProTrek, with its tri-modal contrastive learning paradigm, presents itself as a unique and unprecedented solution capable of charting this complex protein world and opening up new ways of exploration and analysis in numerous disciplines of science.
Methodology Used
The approach used in the study included treating the protein model called ProTrek as the implementation of SaprotHub, which enabled tri-modal alignment training. This is true because the approach used in the training was to use real sample pairs to complete the sequence-structure-function (SSF) tasks. Moreover, the contrastive predictive coding and the alignment with MMseqs2 were performed with only the default settings considering the sequence searching and alignment procedures. The study also uses fine-tuning settings based on SaprotHub guidelines and running benchmarks of alignment-based tools such as Foldseek with basic benchmark tests.
ProTrek Architecture
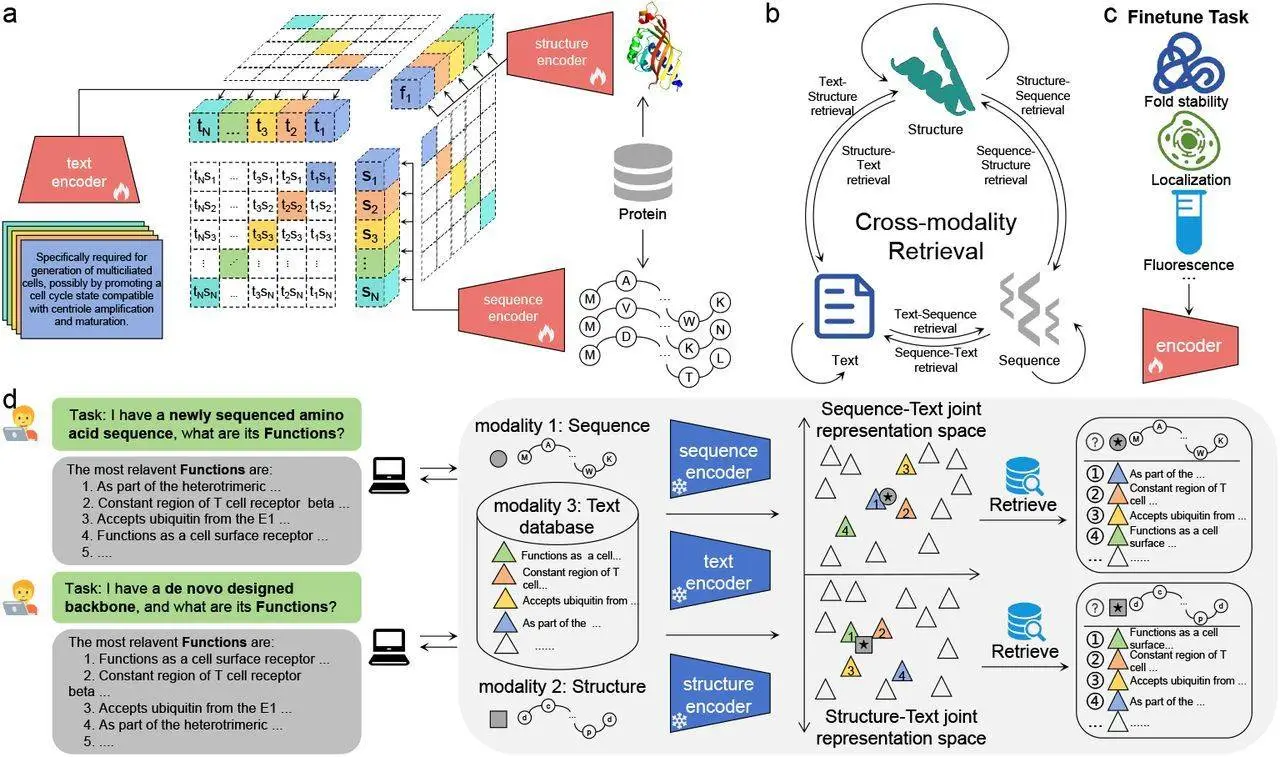
The term used in ProTrek for the search is natural language because it can be used to input any protein sequence and offers maps for easy navigation. Thus, nine search tasks exploring all possibilities essentially of SSF make ProTrek a unique protein representation tool enriched by nine specialties of Trek. Another feature of its contrast learning scenario is its tri-modal approach, which allows the SSF to be associated without a gap by employing supervised transfer learning to improve job prediction.
The Potential of Tri-Modal Contrastive Learning
Here, contrastive learning is employed using Tri-Modal Contrastive Learning to represent a feasible solution to capture the complex interlinkage between protein sequence, structure, and function (SSF). It would be rather instrumental in opening the floodgates of understanding proteins better, one of the basic units of life. ProTrek is a state-of-the-art protein language model that employs the approach of three-modal contrastive learning to the challenges existing in the protein universe.
As a result of employing contrastive learning techniques, ProTrek learns a general-purpose protein embedding that can be more easily adapted to produce usable information on multiple related tasks like fitness and stability prediction. By improving the alignment of SSF through some real sample pairs, the model has the potential for assigning different attributes of proteins appropriately. The tri-modal approach, called “ProTrek,” approaches the study of protein SSF relationships in a manner that is radically different from traditional methods, giving researchers a wealth of knowledge from the start.
This research demonstrates the effectiveness of applying tri-modal contrastive learning in protein analysis in order to view the vast area of proteins that currently remains untapped. In addition to previously existing protein modeling, ProTrek can provide research with many more features that will allow them to explore the paramount aspect of protein structures and functions. As protein research continues to evolve, tri-modal contrastive learning stands out as a promising approach with the potential to revolutionize the field.
Evaluation and Performance
The proposed system ProTrek achieved better results than state-of-the-art methods, such as ProteinDT and ProtST, on the benchmark for search tasks, both for sequence-to-text and text-to-protein retrieval. By training on a relatively large dataset, the present study showed that ProTrek would be an invaluable tool in current protein contexts and bettering current models.
Applications and Implications of ProTrek
Innovative Applications
ProTrek, the near-universal protein model, provides the efficient, large-scale, retrieval-enabled language model to uncover protein sequence-structure-function relationships. It allows the researchers to execute nine different search tasks in the protein universe; it is arguably one of the most powerful search tools today, given insights that it yields on protein sequences, structures, and functions. For this reason, ProTrek is considered an effective tool for protein study and analysis chiefly due to the use of integrated datasets whereby one can easily access protein information contained in massive databases.
Implications for Protein Research
ProTrek provides a universal search and representation method for protein structures with its ideas, which plays a vital role in expanding people’s knowledge about the protein universe. This opens up new possibilities for scientific research in bioinformatics, molecular biology, and protein design, thanks to ProTrek’s capabilities to search across all data modalities and utilize representation models. The generality and the high speed of ProTrek’s search put a spotlight on the role of the program in the development of protein investigation and made a step forward in the sphere.
Conclusion
As for ProTrek, with the functions of cross-modal search and fast running in protein research, representation models ensure that the platform can provide a significant impact in the relevant field. In providing an all-encompassing view of the SSF relationships of proteins, ProTrek opens the door to incorporating complex concepts of protein bioinformatics into the design of proteins, the creation of question-answer contexts, and more. As a proteomic search and representation engine of unmatched dimensions, ProTrek is now all set to chart new frontiers in the prospecting and interrogating the protein realm.
Article Source: Research Paper |Pre-trained ProTrek can be downloaded from the Huggingface| Structural sequences are available at GitHub| Text descriptions are available at Uniprot| Code repository is available GitHub| ProTrek web server is located at Demo_ProTrek
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.