The emergence of Protein Language Models (PLMs) is changing biological research for the better through efficient prediction of functional annotations and protein structures. However, the actual features and mechanisms of these models still largely remain unclear. This posed a critical question that sparked the curiosity of Elana Simon and James Zou, who are both researchers at Stanford University — what PLMs learn, and how can they offer practical assistance to protein biology? Their answer, the InterPLM framework, proposes to take advantage of Sparse Autoencoders (SAE) to interpret features of PLMs and gain a better understanding of the complex information that modern models encode.
However, do not think that this work is simply about the interpretation of the models — it’s about the development of the tools that translate between the languages of AI and biological discoveries. InterPLM serves as a beacon for researchers seeking to understand PLMs and use them to uncover both well-established and novel biological phenomena.
Understanding the Need
The researchers recognize that protein sequences can be interpreted as a ‘biological language’ capable of encoding rules for structure and function. PLMs require copious amounts of protein sequence data for training using transformer-based architectures, with real-world protein sequences performing quite impressively in relevant tasks. While their performance in tasks like protein engineering and annotation prediction is impressive, their methods remain opaque. What exactly do they learn? How are these embeddings, if at all, related to the actual biological features?
One key obstacle lies in the phenomenon of “superposition,” where multiple unrelated biological concepts overlap within the same neurons. This blending obscures interpretability, making it challenging to draw clear connections between the model’s outputs and underlying biology. However, tackling this particular problem requires analysis above the classic neuron level, which is finding a way to disentangle these intricate patterns.
The InterPLM Solution
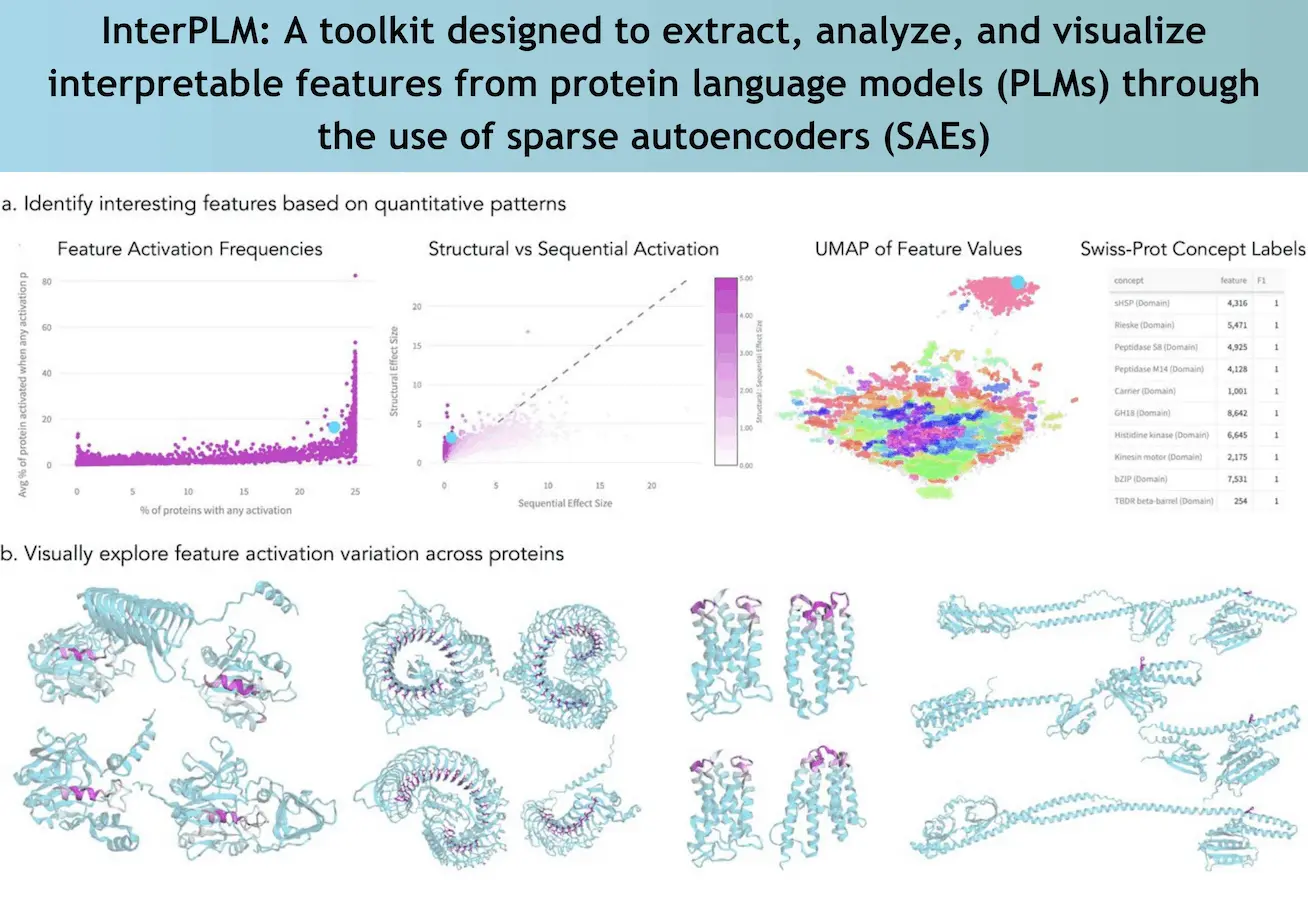
At the heart of the InterPLM framework is the application of Sparse Autoencoders. Unlike raw neuron activations in PLMs, which tend to encode multiple concepts simultaneously, SAEs transform these embeddings into sparse, human-interpretable features. Each feature can be described as encoding a single core biological concept, for example, a structural motif or a binding site; this enhances the interpretability and analysis of the model’s predictions.
Following this approach, the team studied ESM 2, which is one of the many PLMs, and discovered interpretable features numbering more than 2548 per model layer. The aforementioned features are mapped to 143 known concepts in biology, including catalytic sites and functional domains. So many features, and very specific features at that, are impossible to ignore as most neurons for most layers are targeted for further studies and only present a few clear associations.
Remarkably, the framework also revealed patterns that extend beyond existing biological annotations. These novel features hint at uncharted territories in protein biology, offering potential leads for further experimental validation.
Practical Applications and Impact
InterPLM is not just a conceptual improvement –it has real-world applications in protein biology. Refocusing a few features with well-known annotations from Swiss-Prot, a well-established protein database, the authors were able to demonstrate the applicability of the SAEs in filling in the missing annotations. For example, some features pointed out some conserved protein motifs that had not previously been annotated and, therefore, had likely been ignored in annotation due to a lack of comprehension of their biological usage.
The framework also provides the possibility of protein sequence generation in a more specialized manner. By turning on particular features, the researchers were able to change what the model predicted towards the structural or functional elements that were required. For example, features associated with periodic repeats of glycine in collagen-like proteins were manipulated, which evidenced the promise of this strategy in carrying out protein engineering tasks.
In addition, InterPLM develops tools for the scientific community rather than limiting itself to the regulation of analysis. Thanks to their interactive platform InterPLM.ai, researchers can see the learned features relative to the protein structures and sequences to which they correspond. This allows all scientists who have access to this tool to discover new things and promotes communication between scientists and new ideas in this field of study.
Challenges and Future Directions
InterPLM has made remarkable achievements, yet the framework does have its challenges as well. Further refinement will need to be done if the approach is scaled to work with more advanced models, such as AlphaFold, which employs structural predictions. In addition, directing protein sequence generation towards more complex biological architectures is still a work in progress.
An unanswered question is the relevance of such features, some of which do not correspond to existing annotations. It is possible that such features describe new biological landscapes, but testing them requires extensive experimental work.
The team has reported on the limitations of the current practice in terms of metrics for biological interpretability evaluation. Although their approach of associating features with Swiss-Prot annotations rests on a sound premise, the conclusion may be enhanced by seeking such validation in both other experimental approaches and models of structure prediction.
The Road Ahead
InterPLM expands the scope of the current understanding of protein language models and their applications. Simon and Zou have unlocked the core concepts behind PLMs by applying sparse autoencoders accompanied by distinctive visualization tools that enable a better understanding of PLMs and how these concepts can be utilized in practice to tackle real-world problems.
The possibilities are enormous. InterPLM opens new horizons for bioinformatics, from protein databases where there are many unknown regions to the development of more complex proteins. With the maturation of these tools, the opportunity to grasp how these molecules work the reverse PLMs presents the opportunity to turn the mysteries of biology into knowledge that can be applied in practice.
While working with resources such as InterPLM.ai and having open-source code, the researchers have prepared for the next step, taking their results as a basis for further research. By revealing the mysteries of PLMs, InterPLM allows us to imagine the future as where AI stands, where AI helps not only to understand biology but also to create and explore new horizons.
Article Source: Reference Paper | An interactive visualization platform for exploring and analyzing learned PLM features is available on the Website. Code for training and analysis can be found on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.