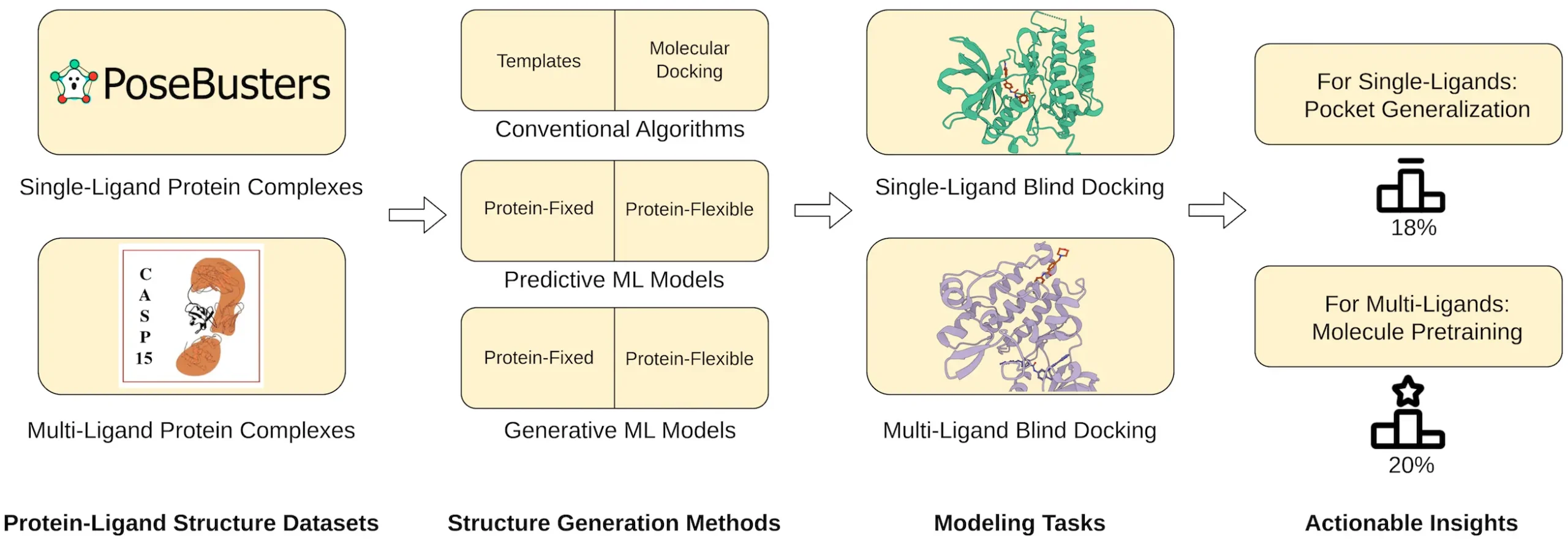
Scientists from the University of Missouri, Columbia introduced PoseBench, the first exhaustive benchmark for practical protein-ligand docking, enabling rigorous evaluation of deep learning docking methods on predicted protein structures, multiple concurrently binding ligands, and targets without known binding pockets. It introduces multi-ligand benchmark datasets and reveals limitations of current deep learning approaches in generalizing to multi-ligand scenarios.
Introduction
The human body is composed of a complex symphony of interactions, where trillions of proteins are involved in every cellular process. But what if we can intervene at the molecular level by introducing little agents to push protein function back into healing? This is the ambitious goal of drug discovery – a relentless pursuit for molecules that can fit into protein structures like missing puzzle pieces, thereby reestablishing harmony and favoring recovery.
However, finding these “magic bullets” is no walk in the park. More often than not, intuitive reasoning, trial and error, and sometimes sheer luck have been used as guides throughout this process. Nonetheless, a new tool has recently emerged: artificial intelligence (AI), more specifically known as deep learning (DL). Scientists now have an entirely new world before them, using AI to dock proteins with ligands during drug discovery.
But are we there yet? Can artificial intelligence solve the riddle behind life and improve the pharmaceutical industry? Recent research by the University of Columbia, researchers explores this very question. They have come up with a revolutionary way to assess how successful deep learning is at protein-ligand docking called POSEBENCH. It presents an interesting peek into AI’s current powers in this field as well as its limitations, thus preparing the ground for future progressions.
POSEBENCH Power
What makes makes POSEBENCH unique?
Realistic Scenarios: Unlike the previous benchmarks, POSEBENCH contains real-life complexities. It tests methods using predicted protein structures that simulate what scientists may meet in their work. Furthermore, it assesses approaches in multiple ligands bound to a single receptor, which is more complex but closer to reality.
Focus on Multi-Ligand Docking: In the past, protein-ligand docking has been mainly concerned with the binding of single ligands. On the other hand, multi-ligand docking has been given a special place by POSEBENCH as its importance grows due to implications for drug discovery.
The Verdict: Promising Yet Room for Improvement
POSEBENCH results give an interesting image. A promising performance was shown by DL methods such as DiffDock-L and DynamicBind in single-ligand docking, but they stumbled on multi-ligand circumstances. Interestingly, some traditional template-based docking algorithms performed better or at par with current DL methods when it comes to multi-ligand docking.
These findings imply that current DL approaches might need to capture the complexity of multiple-ligand interactions fully. The molecule pre-training process is emphasized in this study, where one trains a model on a large range of molecular data before fine-tuning it for protein-ligand docking. Through pre-training, models could learn to understand various interactions between different molecules, hence gaining more knowledge on how to handle multiple ligands during their docking in proteins.
The Road Ahead: Defining the Locks
Further development of DL methods for protein-ligand docking, especially in multi-ligand scenarios, is necessary, as highlighted by the POSEBENCH study. Some appealing suggestions for future research are mentioned below:
- Multi-Ligand Focused Training: Researchers could improve their performance in this area if these models were created specifically for multi-ligand docking datasets.
- Leveraging Molecule Pre-Training: It is highly promising to consider whether fine-tuning DL models on various molecular data sets after pre-training them with diverse molecules can be effective for protein-ligand docking.
- Incorporating Biophysics: Incorporating principles from biophysics into the training process of DL models has the potential to allow DL models to simulate real protein-ligand interactions and predict steric clashes (places where two molecules occupy the same space).
Conclusion: The Future of Drug Discovery, Unmasked Together
The POSEBENCH study is a stepping stone towards the full potential of AI-powered molecular docking. As scientists continue to develop DL techniques and work more extensively on multi-ligand interactions, we expect considerable advances in drug discovery. Imagine a future where personalized medicines are designed with unparalleled precision, targeting specific protein interactions and leading to more effective treatments with fewer side effects.
We do not need to worry; the future will be bright if we maintain our present efforts at collaboration among computer scientists, biochemists, and pharmacologists.
Join the Conversation!
What are your thoughts on the future of AI applications in drug discovery? Can DL methods go beyond traditional docking approaches? How do we bridge the gap between computational modeling and real-world drug development?
Feel free to share your thoughts and ask questions in the comment section below! By maintaining an open conversation, we can hasten to create new solutions that will unlock many mysteries behind protein-ligand docking for a healthier tomorrow.
Article Source: Reference Paper | POSEBENCH code, data, tutorials, and benchmark results are available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.