Proteins are life’s molecular workhorses and play crucial roles in every biological process. Understandably, their complex shapes, also known as conformations, are the key that unlocks their secrets to new drug development. Proteins are not still statues but rather change into different conformations like ninjas to play different functions. This movement of protein has been a major challenge during drug discovery because traditional methods usually capture a single snapshot of a protein.
A glimmer of hope comes from a recent study published in Nature Communications by a team at Brown University. Their investigation introduces a brand new method that employs artificial intelligence (AI) to predict several shapes that proteins can adopt. If it is upheld, this advancement could alter our perception of protein functioning and lead to better medications.
The Challenge: Beyond the Static Picture
As stated by the media release on the Brown University website, protein structures were conventionally identified using X-ray crystallography, among other techniques. Despite their value, however, these methods give a static image, just like a still photo of protein in one pose. Nonetheless, proteins are dynamic entities that change their shapes continuously to perform their function. This dynamic nature is important for understanding how drugs interact with proteins. A protein could be likened to a lock while the drug is seen as the key; if the protein changes shape, then the key may not fit any longer, and hence, this renders it useless for what it was meant for.
AlphaFold2: Powerful Yet Constrained Tool
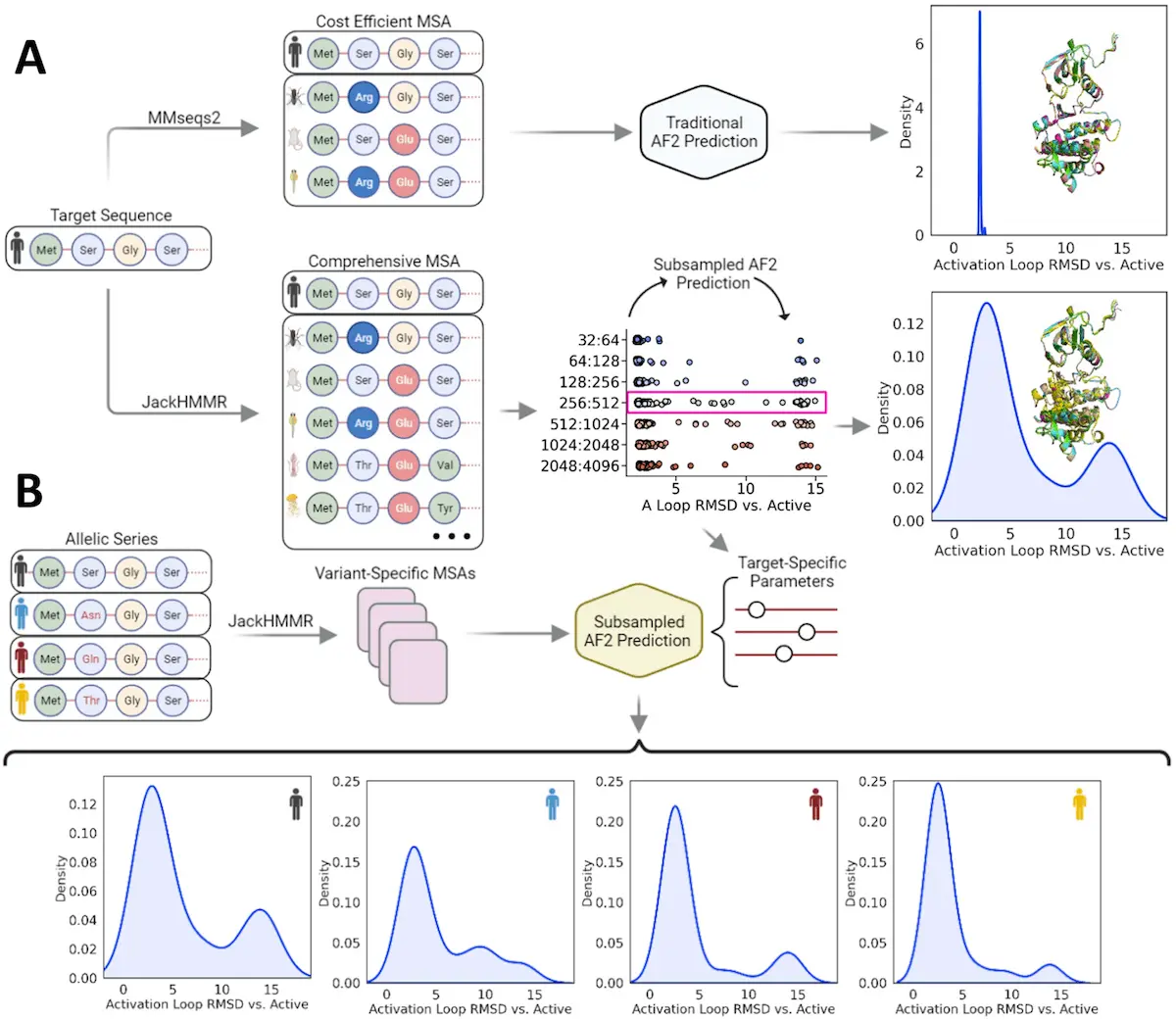
AlphaFold2, developed by DeepMind, has been a significant advancement in protein structure prediction. This AI-based approach can accurately predict the most stable conformation of a protein. However, according to Monteiro da Silva et al., AlphaFold2 has one serious drawback—it doesn’t predict all possible shapes of the protein. It’s like having an HD picture of one key, but you don’t know any of its locks.
Second Inventory: Subsampled AlphaFold2 Changes Everything
The Brown University scientists tried thinking out of the box about overcoming limitations associated with AlphaFold2 predictions. They argued that perhaps the information necessary for predicting a protein’s conformational flexibility lies within its amino acid sequence. These methods, although useful, give a static image of it, like a photograph of a protein that has been frozen in just one position. Nonetheless, proteins are dynamic creatures and keep changing their shapes to carry out their tasks. This dynamic attribute is important for knowing how drugs influence proteins. If you think of the drug as being the key and the protein as a lock, then a slight change in the shape of these locks may make it impossible for this key to fit anymore.
Proteins undergo mutations that change the sequence of their amino acids and alter their structure. They believed that by finding out its evolutionary path through its amino acid sequencing, some light would be shed on its mutability.
Furthermore, there’s what their technique does. To AlphaFold2, they provided “subsampled” input data. Think about delivering AlphaFold2 with an entire picture regarding a protein sequence but not exactly doing what they had to do. Nevertheless, instead of providing it with all parts of a sequence, they gave it small portions, hoping that this would confuse it and allow it to explore more possible conformations. Many might have thought that such an idea could not work, but this study provides evidence that this approach is plausible. For instance, when subsampled, Alphafold2 predicted two test proteins’ various structures very accurately, even indicating slight changes in shape changes resulting from mutation.
Drug Discovery Implications: A Better Tomorrow?
The capacity to predict the conformational landscape of a protein is a game-changer for drug discovery. Drugs are made to work on particular protein shapes. By fully comprehending protein dynamics, scientists can create drugs that more easily bind to active conformations of proteins, thus giving them a more powerful and specific therapeutic effect. Moreover, it could identify new drug targets – proteins that may have been neglected because they did not seem like a good target in their static conformation.
Looking Forward: Beyond Validation
This study represents a major advance, but further validation is required. The researchers validated these predictions using experimental data from other techniques, such as nuclear magnetic resonance for the two test proteins. However, this may only be possible across some proteins. More work needs to be done so that independent methods can be developed to validate the accuracy of AlphaFold2’s subsampled predictions.
However, validating the accuracy of subsampled AlphaFold2 is just the beginning of another journey altogether. Researchers have been working on improving the technique and expanding its capabilities into other areas. These include:
Supplementary Data Integration: At present, only the protein sequence information is used in subsampled AlphaFold2. For instance, consider a protein’s surroundings, such as other molecules, or experimental findings from X-ray crystallography. This extra data could help refine the predictions further and account for complex variables affecting the shape of proteins.
Bypassing Prediction: Modelling Dynamics: While predicting stationary poses is a worthwhile endeavor, it should ultimately be able to capture all motions that a protein carries out. Some researchers have proposed that sub-sampled AlphaFold2 be combined with molecular dynamics simulations. They might lead to a better understanding of how proteins stretch and bend.
Automating the process: Currently, this process of data subsampling relies upon human intervention. The future direction in this field is automating this procedure, which would enable large-scale analysis of protein dynamics across many datasets. This could be helpful for those who are working on protein families or trying to find out what happens when multiple mutations occur.
While this is interesting, it must also be recognized that there are limitations to it. For example, highly flexible proteins may not have all their possible conformations predicted by the sub-sampled AlphaFold2 technique. Additionally, the accuracy of the predictions might depend on the quality and completeness of the protein sequence data.
The Democratization of Drug Discovery
Drug discovery has been traditionally known as a time-consuming, costly process. If confirmed and advanced, Subsampled AlphaFold2 has the potential to democratize this process. Here’s how:
Reduced Costs: Predicting protein dynamics through subsampled AlphaFold2 may be much cheaper and faster than traditional methods. This could offer opportunities for smaller research labs as well as academic institutions to join drug discovery efforts.
Repurposing Existing Drugs: Using this technique would identify ways in which existing drugs interact with different protein foldings and discover new uses for them. This can assist in the quick development of new therapies for existing diseases.
Personalized Medicine: A reality can be made of personalized medicine by understanding protein dynamics within an individual’s specific genetic context. Just think about drugs that are designed to strike specific conformations of proteins found within the body of a given patient.
Conclusion: A Promising Path Forward
Monteiro da Silva et al.’s recent method is a significant tool for investigating protein motion. It can completely transform our comprehension regarding how proteins work and, therefore, provide better medicines that we have been waiting for all along. More research is necessary to prove and assert its correctness and deal with the restrictions thereby. The way to a breakthrough in science starts with a promising technique and leads to a revolutionary tool, but it needs careful assessment and adjustment. However, due to the imaginative manners of scientists, there is hope for a better future in terms of protein dynamics and drug discovery.
Prospects of protein dynamics are immense for medicine revolutionization and understanding of life in general. New therapies that target diseases could be ushered through employing novel methods such as subsampled Alphafold2 in conjunction with coordinated trials that unlock the structural secrets of these dynamic molecules to yield a greater understanding of life’s changing nature.
Article source: Reference Paper | Reference Article | Scripts used in the study are available for public use on GitHub
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.
















[…] Brown University’s Subsampled AlphaFold2 Approach for Predicting Protein Dynamics: A Game-chan… […]