The synergy among the Nuffield Department of Medicine and the Department of Statistics at the University of Oxford represents a blend of medical expertise with statistical rigor – “The MolSnapper,” built using deep learning, promptly exploring intricate molecular specificity for proteins in 3D conformations. Unlike previous approaches that rely on costly trial and error, Conditioning Diffusion uses deep learning to navigate the maze of molecular interactions with unprecedented efficiency to provide novel medical research solutions.
Breaking Down the Fence
Their research bridges the gap by making it less complicated to evolve patterns learned from large molecular datasets for pocket-binding applications. MolSnapper employs a controlled and environmentally friendly producing approach to seamlessly combine 3-D structural insights with expert knowledge, revolutionizing the ligand era in pharmaceutical applications. MolSnapper enables researchers to generate molecules from large datasets by providing personalized instruction and enhanced selection. This differs from conventional structure-based drug design approaches, which are stereotypically proficient on restricted protein molecular data.
Ability of Precision
Precision is imperative in drug design and discovery. Therefore, MolSnapper authenticates this concept by serving to deliver personalized descriptions specific to the investigator’s requirements. With the potential for drug discovery employing deep learning, the path has been opened for personalized medicine intensive on molecular markers unique to particular diseases.
Evaluation of MolSnapper to Other Approaches
To affirm the efficiency of MolSnapper in conditioning ligand generation inside a protein pocket, researchers performed massive comparisons with contemporary procedures designed for analogous reasons along with the following:
- MolDiff was used to evaluate the efficacy of the conditioning method, and they compared MolSnapper to it, a way of producing ligands without conditioning. Following MolDiff molecule synthesis, the Vina method was used to dock the subsequent ligands. This step was crucial because MolDiff-generated molecules no longer fit the pocket.
- SILVR (Selective Iterative Latent Variable Refinement), a technique that generates compounds optimized for protein-binding sites, was used to define pharmacophore positions and atoms as references and contrary atoms as dummy atoms. The SILVR rate is 0.01, consistent with the approach defined in the original reports.
- DiffSBDD uses Inpainting, a method for fixing simple molecular structures. In their DiffSBDD experiments, pharmacophore positions and atoms have been used as inputs for constant atoms. For every experiment, 100 new ligands were synthesized for each context, with three tries in step with generation to ensure a radical assessment of ordinary overall performance and reliability.
Experimental Arrangement for Employing MolSnapper in Structure-Based Drug Discovery
The investigational setup for the MolSnapper in structure-based drug finding comprises several vital components:
Dataset Assortment: Researchers acknowledged data sets comprising CrossDocked2020 and Binding MOAD, which incorporate experimentally determined protein-ligand complexes. These datasets are curated in line with exact criteria, along with passing all PoseBusters valuations and having a low number of hydrogen bond interactions.
Preprocessing: The primary dataset of protein-ligand complexes is preprocessed to authorize its gratification and significance. This entails complexes that do not fulfill the given standards and indicating examples with critical pharmacophores and hydrogen bond relations.
Model Training & Conditioning: MolSnapper makes use of a generative diffusion version referred to as MolDiff, which turned into a pre-trained procedure of the GEOM drug dataset. The model is conditioned without retraining, which includes expert information through 3-D pharmacophores to strain chemical advent optimized for precise protein interaction.
PoseBusters Test Suite: MolSnapper’s overall performance is measured by using a lot of metrics, cooperatively with structural and chemical similarity to reference molecules, the presence of hydrogen bond interactions among ligands and proteins, and the functionality to generate molecules that pass PoseBusters endorsement and consistency assessments.
Comparison with Substitute Methods: Researchers designed MolSnapper following the aspects of methods such as SILVR and DiffSBDD to boost the performance of designed molecules with greater binding affinity and selectivity, much like reference compounds.
Ablation Studies: Ablation tests are used to explore the impact of numerous initiation approaches and reference entity treatments on molecule formation. This allows us to apprehend the location of noise in molecule formation employing graph neural networks (GNNs).
Concluding Remarks
MolSnapper is a unique method for enhancing diffusion fashions with the aid of the usage of collectively with 3-D pharmacophoric limitations. This groundbreaking technique enables genuine molecular generation, allowing users to convert beyond records into extra suitable molecule design processes. MolSnapper outpaces prior techniques, including SILVR and DiffSBDD, with up to a threefold growth inside the sort of molecules that bypass PoseBusters tests. Additionally, it improves form and coloration similar to reference ligands by up to 20%, resulting in a 30% increase in first-hit retrieval.
Article source: Reference Paper | Source code available at GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.

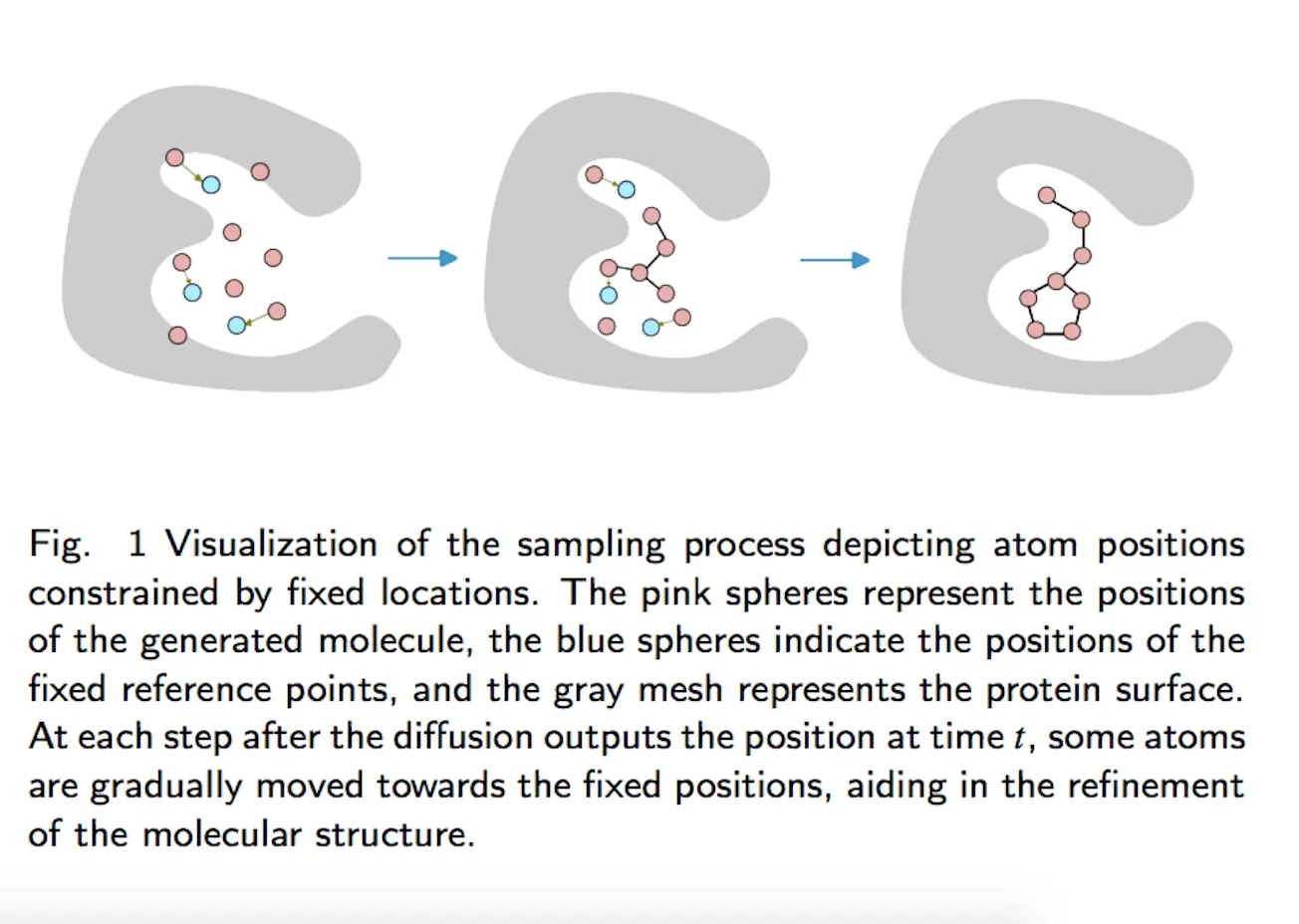















[…] Bridging Gaps in Structure-Based Drug Design: The MolSnapper’s Breakthrough Conditioning Diffu… […]