Researchers at the University of Singapore introduced “Melvin,” an Alexa Skill that lets you query cancer genomics data with your voice. This groundbreaking tool bypasses complex interfaces and empowers anyone to ask questions about massive cancer datasets through simple conversations. Melvin promises to democratize cancer research and personalized medicine by putting this life-saving information at everyone’s fingertips.
While extensive cancer genomics data sits within reach, its exploration remains sluggish for countless researchers and clinicians, hindering progress in the fight against the disease. Existing tools like Graphical user interfaces (GUIs) for exploring data sets like The Cancer Genome Atlas (TCGA), though well-intentioned, feel lagging and opaque, leaving users longing for a faster, more intuitive way to glean insights.
However, modern interfaces leveraging augmented intelligence have the potential to streamline data exploration in cancer genomics. In particular, voice user interfaces (VUIs) can enable users to query complex data quickly using natural language. A new study published in Nature Communications Biology introduces “Melvin,” an Alexa skill that unlocks the power of massive cancer genomics datasets through simple voice conversations. By bypassing the limitations of traditional interfaces, Melvin empowers anyone to ask crucial questions about this life-saving information, potentially accelerating research. This voice-powered tool could democratize data analysis and transform the future of cancer care.
Melvin’s Cancer Genomics Knowledge Base
Melvin contains harmonized genomic data sets representing all 33 TCGA cancer types. Users can inquire about somatic mutations, copy number alterations like amplifications and deletions, and gene expression levels. As a proof-of-concept, the researchers also integrated mutation and copy number data from the Breast Cancer Somatic Genetics Study (BASIS) to demonstrate Melvin’s ability to incorporate additional data sets.
Ancillary information like gene definitions and therapeutic actionability data is available to help contextualize and interpret results. Importantly, Melvin can email all results and visualizations to users instantly.
Intuitive Voice-Based Analytics via a Finite State Machine
Asking complex questions about cancer genomics data can be difficult to verbalize in one sentence. To overcome this, Melvin allows users to provide query attributes incrementally in any order.
This multi-turn conversational process uses a finite state machine framework, where Melvin provides relevant biological responses based on the current conversation state. This retains context to enable rapid follow-up questions without repeating previous attributes.
For example, after querying TP53 mutations in breast cancer, users can simply say, “How about PIK3CA?” to switch genes. This rapid navigation allows seamless exploration across genes, cancer types, and data types.
Powerful Comparative Analytics Through Voice
Melvin’s state-based architecture also enables more advanced analytics using natural language. The compare feature contrasts two values of the same attribute type given the current context.
Users can easily assess the co-occurrence of alterations, compare frequencies, and juxtapose putative driver genes between cancer types conversationally. The split-by function determines how a quantitative variable (like gene expression) varies based on a binary variable (e.g., mutation status).
This demonstrates how Melvin automatically maps conversational interactions to analytical pipelines based on the conversation history and current state.
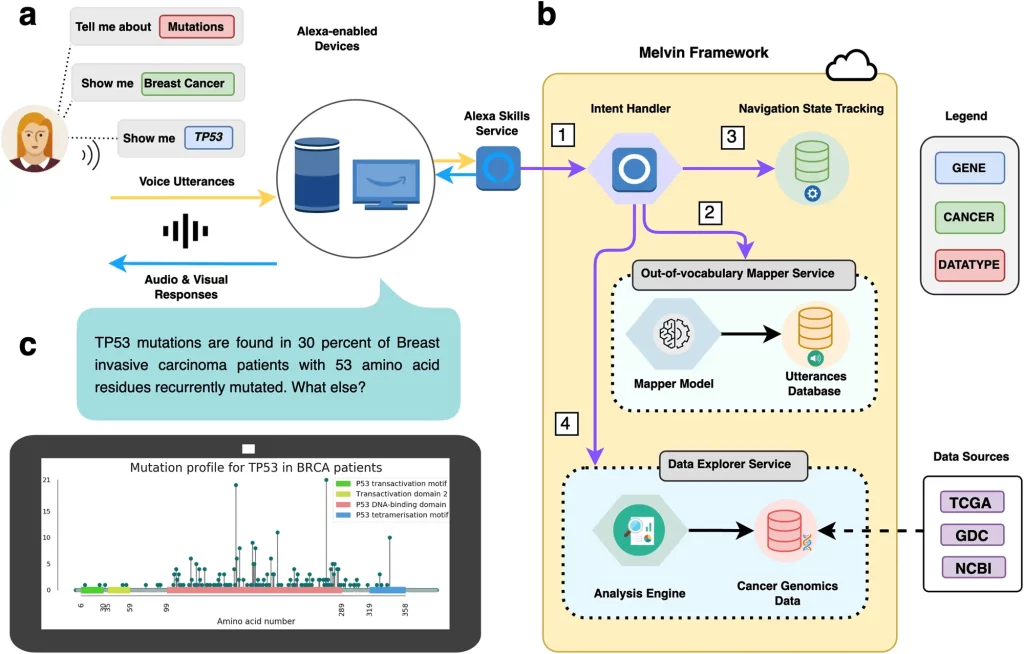
Image Source: https://doi.org/10.1038/s42003-023-05688-z
Crowdsourcing Utterances to Improve Gene Name Recognition
Correctly interpreting users’ natural language requests is critical for Melvin’s functionality. The researchers developed an approach to crowdsourcing pronunciations for cancer-associated genes, cancer types, and data types from domain experts.
This dataset of over 24,000 utterances helps power Melvin’s out-of-vocabulary mapper service (OOVMS) to improve speech recognition and entity mapping. When Melvin cannot map a user’s speech to a known attribute, a classifier predicts the best match based on trained utterance features.
Overall, Melvin’s OOVMS correctly resolved 88.9% of test utterances, a 69.8% boost over Amazon Alexa’s baseline performance.
Allowing Users to Specify Personal Pronunciations
To further improve accuracy, a web portal is built where users can submit personalized pronunciations for genes of interest. Users record their own audio pronunciation, which teaches Melvin to map that vocalization to the intended gene.
These personal utterances integrate with users’ Alexa profiles to progressively enhance recognition. This underscores Melvin’s ability to improve through crowdsourcing and user feedback.
Conversational Interfaces to Democratize Cancer Genomics
Melvin demonstrates the potential of voice interfaces for efficient cancer genomics data exploration. By enabling natural language queries, Melvin facilitates democratized analytics without deep technical expertise.
As voice technology matures, conversational agents like Melvin could assist clinicians in interpreting panel sequencing reports within molecular tumor boards. Hybrid platforms integrating voice, databases, and large language models may further enhance these applications.
While Melvin currently focuses on TCGA, the researchers continue to add valuable cancer genomics data sets over time. Melvin’s codebase is open-source to spur community-driven extensions as well. Overall, by simplifying access to cancer genomics resources, Melvin exemplifies how conversational AI can empower scientific discovery and clinical translation.
Conclusion
Melvin provides an intuitive way for researchers, clinicians, and the general public to explore and analyze cancer genomics data conversationally. By retaining context across multi-turn interactions, Melvin allows both simple and complex queries to be resolved quickly through natural language.
Crowdsourcing pronunciations combined with personalization improves Melvin’s voice recognition capabilities over time. As cancer genomics databases continue expanding, innovations like Melvin will be critical to extracting key insights and making these resources more accessible. Conversational interfaces represent a promising approach to democratizing cancer genomics and augmenting scientific inquiry moving forward.
Story Source: Reference Paper | Website | Melvin is freely available as an Amazon Alexa skill (https://www.amazon.com/dp/B09NZSRBNS).
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
















[…] Alexa, Analyze Cancer Data: Meet Melvin, Your Virtual Genomics Assistant […]
[…] Alexa, Analyze Cancer Data: Meet Melvin, Your Virtual Genomics Assistant […]