Large-scale screening techniques that are resource-intensive and offer limited control on binding mode are usually used to produce macrocyclic binders to therapeutic proteins. There are currently no reliable methods for the de novo design of protein-binding macrocycles, despite significant advancements in deep-learning methods for protein design and physics-based methods for peptide design. In this work, scientists from the University of Washington present RFpeptides, a denoising diffusion-based pathway for creating macrocyclic peptide binders that target specific protein targets. A sub-nanomolar IC50 and a KD of 6 nM have been established for the anti-GABARAP macrocycle design models. The target, RbtA, does not have an experimentally defined target structure, but it possesses a high-affinity binder with KD < 10 nM. Twenty or fewer developed macrocycles were tested against four different proteins in a study, and the results showed medium to high-affinity binders against all of the chosen targets. This design overcomes the drawbacks of library screening techniques by offering a strong framework for the quick and personalized design of macrocyclic peptides for therapeutic and diagnostic uses.
Challenges and Opportunities in Developing Macrocyclic Peptide Therapeutics
Macrocyclic peptides present a possible path for the creation of novel treatments that close the gap between big biologics and small-molecule medications. A novel therapeutic strategy is offered by these peptides’ ability to modify molecular targets that are unavailable to conventional therapeutic techniques. Although they can reach intracellular targets, small compounds are not the best choice for targeting proteins that don’t have deep hydrophobic pockets.
Peptide therapies have historically depended on either high-throughput screening of trillions of random peptides for target binding utilizing display-based methods or natural product discovery. However, these approaches have drawbacks, such as low mutational tolerance, marginal stability, and synthetic difficulties. Due to the exact structural control needed to attain these functional features, high-throughput screening techniques are labor-, time- and cost-intensive and frequently fall short of optimizing for a variety of biophysical parameters, including target binding, selectivity, and membrane permeability.
Looking into Previous Studies
Structure-guided design methods offer an alternative to library screening and allow scientists to quickly explore a wide range of chemical and structural varieties while building macrocycle binders for therapeutic targets. These techniques were created utilizing physics-based methodologies and have demonstrated excellent accuracy in the design of structured macrocycles, binders to protein targets, and hyperstable-restricted peptides. However, the design of protein-binding macrocycles has not been very successful; sometimes, they do not agree with the design models and only obtain modest binding affinities. Additionally, these methods are limited to well-researched protein targets due to their dependence on already-named binding partners.
Promising deep learning techniques have been reported to anticipate macrocycle structures and develop peptide binders to protein targets. Recent work has revealed a pipeline for hallucinating and predicting the structures of macrocyclic peptide monomers. However, neither the broad structural validation nor the ability to undertake atomically accurate de novo creation of macrocyclic peptide structures in complexes with a variety of protein targets has been demonstrated for these methods.
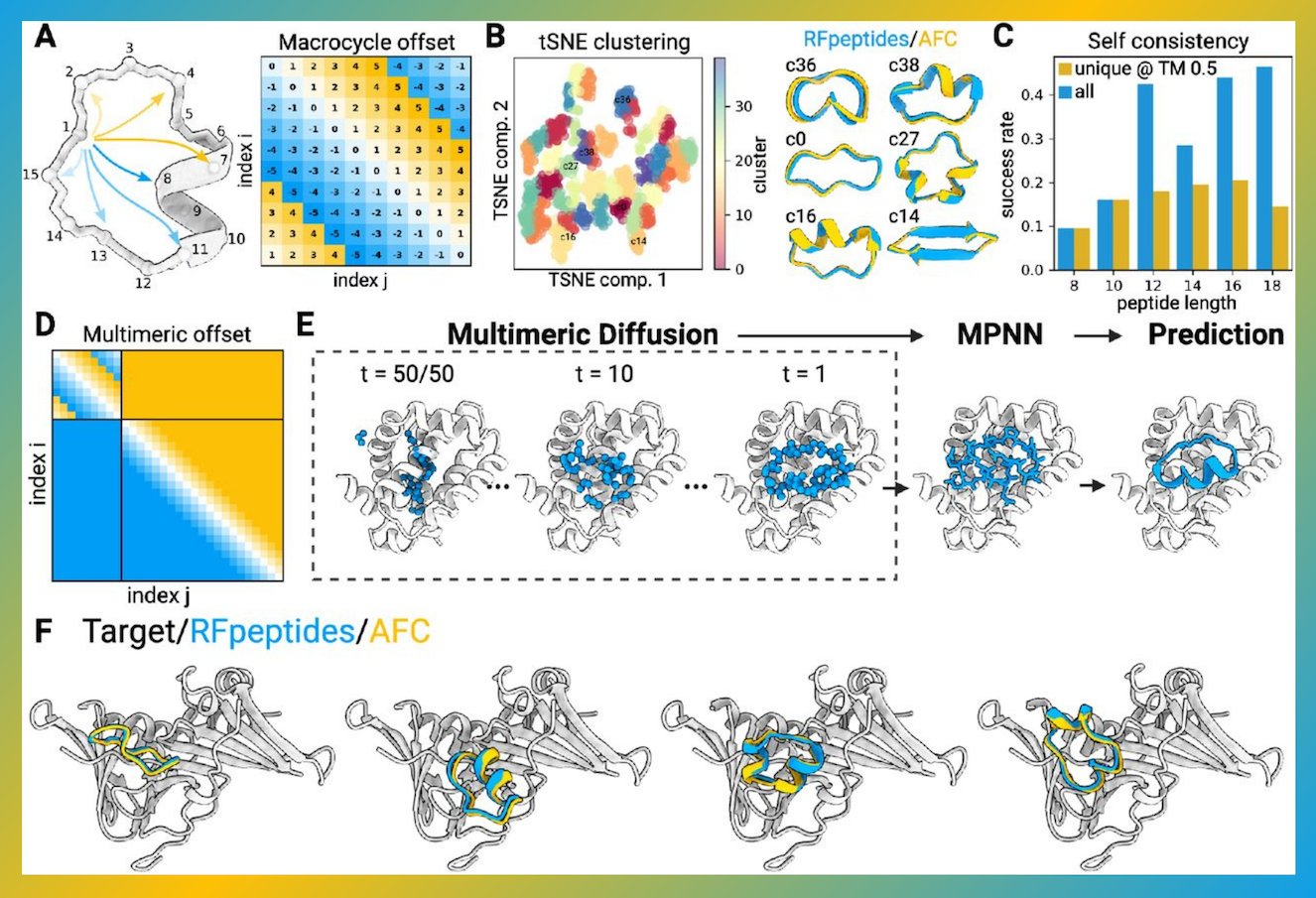
Understanding RFpeptides
A generative deep learning pipeline called RFpeptides is used to precisely construct macrocycle binders with respect to a variety of protein targets. The nearly identical X-ray crystal structures and design models of the macrocycle-bound MCL1, GABARAP, and RbtA (Cα RMSDs of 0.7 Å, 1.2 Å, and 1.4 Å, respectively) illustrate the approach’s strength, as do the high affinities (Kd < 10 nM) of the designed macrocyclic binders to GABARAP and RbtA. By creating high-affinity binders for two targets without additional experimental tuning, the RFpeptides methodology provides a more effective target binding design approach with a greater success rate than any other method.
Custom binders to particular patches and sites are possible with this design technique, which gets around the drawback of complicated structure determination in conventional library-based systems. Because the design models are atomically accurate, the bottleneck of complex structure determination can be avoided using structure-guided optimization for qualities other than target binding. Beyond the constraints of conventional techniques, RFpeptides may make it possible to build peptides that are simultaneously optimized for target binding, cell permeability, or oral bioavailability when combined with membrane traversal design principles.
Conclusion
The ability to create macrocycles de novo from the target’s structure or sequence gives RFpeptides a major edge over earlier computational peptide design techniques. This allows for design against molecular targets that were impossible to target with earlier techniques. To pick topologies that are suitable for the protein they are targeting, RFpeptides create macrocycles of various sizes and forms. The four targets that were evaluated were as follows: binders for MCL1 and MDM2 have helical motifs, binders for GABARAP have a β-sheet topology, and binders for RbtA sample loop-like topologies make extensive contacts with this target’s flat surface. Using this method speeds up the process of creating peptides for various functional uses by allowing the quick design of unique macrocyclic binders that can bind to a broad range of molecular targets. This design offers a strong foundation for the fast and personalized creation of macrocyclic peptides for therapeutic and diagnostic uses.
Article Source: Reference Paper | The RFpeptides pipeline code and scripts will be released by the researchers upon publication.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.