Researchers at the University of Pennsylvania developed PeptiVerse, a single platform that predicts drug-related properties of therapeutic peptides using either amino acid sequences or chemically modified SMILES representations. The platform uses pretrained protein and chemical language models to make quick and precise predictions for various peptide properties. With a web-based interface and an open-source implementation, PeptiVerse allows users to easily evaluate, compare, and optimize peptide candidates. This supports early-stage drug development and peptide design focused on properties.
Peptides as emerging therapeutic agents
Peptide – a short chain of amino acids linked by a peptide bond, peptides have been gaining their place as therapeutics. GLP-1 peptide drugs which are currently frequently used to treat obesity and diabetes. Peptide has advantages over small molecules since they can bind to large protein surfaces that are typically inaccessible to small molecules. In addition, they are simpler and smaller than antibodies, so they reduce the number of immunological responses and are easier to manufacture.
Despite being promising drug molecules, natural peptides frequently have low solubility, rapid enzymatic degradation, poor cell entry, and low oral availability, all of which can limit their efficacy. Chemical changes like cyclization or the use of artificial amino acids can address these problems. However, when the peptides are chemically modified, it becomes difficult to predict their properties or accurately analyse them with existing tools available.
For the design of new peptide drugs, current computational tools are insufficient. While small-molecule ADMET tools accept SMILES inputs but are trained on drug-like chemicals rather than peptides, sequence-based models such as PeptideBERT only work with natural amino acids and cannot handle chemical modifications. Mutation-focused platforms do not evaluate crucial drug-relevant characteristics, and some peptide-specific SMILES tools take chemical structure into account but only cover a handful of properties. All things considered, there isn’t a single adaptable tool that can assess all significant therapeutic characteristics of both naturally occurring and chemically modified peptides.
Model Architecture of PeptiVerse
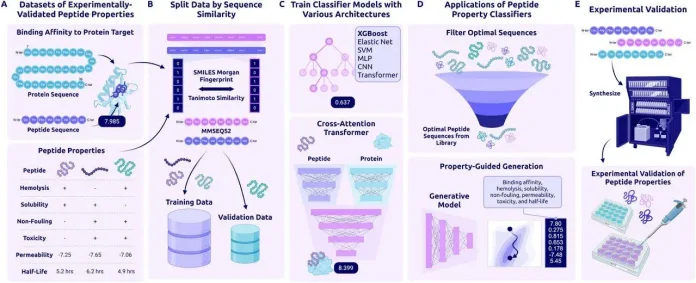
PeptiVerse is a comprehensive platform for evaluating drug-like properties of therapeutic peptides. It supports both canonical amino acid sequences and chemically modified peptides represented as SMILES and uses advanced AI models to produce quick and precise predictions.
PeptiVerse is a tool for peptide drug development that assesses peptide properties and provides generative design workflows to help prioritize and optimize candidates. It encodes amino acid sequences using the ESM-2 model and extracts non-natural peptide chemistry from SMILES inputs using PeptideCLM. Embeddings are either averaged to represent the peptide as a whole or position-specific to preserve residue-level information, depending on the prediction task.
Permeability, solubility, toxicity, hemolysis, non-fouling behavior, binding affinity, and half-life are some of the many characteristics that PeptiVerse predicts. It uses both conventional machine learning techniques (SVM, Elastic Net, and XGBoost) and deep learning models (MLPs, CNNs, and transformers). To guarantee accurate and repeatable predictions, Optuna is used to optimize the hyperparameters for every model, and each model is trained in its typical configuration.
PeptiVerse accurately predicts the strength of peptide–protein binding, in comparison with structure-based methods like ipTM that have trouble with flexible or chemically modified peptides. It is useful for peptide design and filtering. The model does not compress the peptide into a single average value. Instead, it keeps information about each amino acid separately, which helps it predict peptide behavior more accurately and match real experimental results for both normal and chemically modified peptides.
PeptiVerse performs on equal levels with or better than current benchmarks when compared to tools like PepLand and PeptideBERT, which are restricted to particular properties or single input types. Its main benefits are a unified multimodal framework for property prediction, realistic similarity-based data splits, and support for both canonical and non-canonical peptides.
Researchers can use PeptiVerse’s user-friendly web interface to predict various peptide properties by entering SMILES strings or amino acid sequences. Additionally, users can compare predictions with experimental results by using the platform’s training data visualizations. PeptiVerse is simple to replicate and incorporate into peptide design workflows because all datasets are publicly available and standardized.
Conclusion
PeptiVerse is a unified platform that predicts various peptide properties for both SMILES-based (chemically modified) peptides and amino acid sequences. It is effective, scalable, and simple to implement because it makes use of pretrained protein (ESM-2) and chemical (PeptideCLM) embeddings with lightweight predictor models.
PeptiVerse demonstrates that simple classifiers with strong embeddings are often sufficient and can perform better than complex SMILES-based models like PepLand, particularly for binding affinity prediction, where structure-based approaches alone fall short. Prediction of numerical values like half-life and binding affinity has smaller and variable datasets, whereas classification tasks like hemolysis, solubility, and non-fouling have large, well-balanced datasets. This highlights the need for property-specific modeling and more experimental data by demonstrating that prediction accuracy is more dependent on data size and quality than on the model itself, effective embeddings and data volume are more important for performance than model type.
The platform is open, extensible, and regularly updated, allowing new data and models to be seamlessly integrated. However, sparse data still limits some tasks, such as predicting half-life for chemically modified peptides. Specialized predictors for metal-binding, motif recognition, and peptide isoform specificity will be included in future updates. Enhancing its potential for therapeutic development and generative peptide design.
Article Source: Reference Paper | Availability: Hugging Face.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Jainab Shaikh is a postgraduate in Biotechnology with a strong interest in understanding how research translates into real-world innovation. Her areas of focus include biosensors, bioinformatics, and sustainable biotechnological applications. She is passionate about exploring recent scientific advancements and communicating them through clear, engaging, and accessible content. Her work particularly emphasizes research-driven narratives in healthcare, biotechnology, skincare science, and emerging life science innovations.















