A new precision medicine tool 'iSNP' uncovers hidden genetic links that could enhance IBD and other difficult illnesses treatment.
A group of scientists and clinicians from the Quadram Institute, the Earlham Institute, the Norfolk and Norwich University Hospital, and the University of East Anglia with collaborators in Cambridge, London and Leuven (Belgium) developed a systems genomics approach ‘iSNP’ to reveal the patient-specific pathogenic pathways and proteins involved in ulcerative colitis (UC).
Identifying which of several possible disease pathways a patient is using, a novel and powerful systems genomics approach will lead to more effective diagnosis and treatments, as well as a much better understanding of this complicated condition, which can be applied to other perplexing disease studies to help more patients.
The workflow’s ability to identify cohorts of disease associated mutations and pathways isn’t limited to IBD; it has the potential for use in other complex disease including mental health, heart disease and autoimmune conditions. To develop precision therapies based on patient specific genetics opens up the possibility of much needed personalised medicine approaches to tackle these complex and poorly understood conditions.
Dr Tamas Korcsmaros, co-lead author of the study from the Earlham Institute and the Quadram Institute.
In the study, the scientists define a precision medicine workflow, the integrated single nucleotide polymorphism network platform (iSNP), intended to determine the mechanisms by which SNPs influence cellular regulatory networks and how SNP co-occurrences add to disease pathogenesis in ulcerative colitis (UC).
In the post-genomic era, identifying key genetic changes for individual patients has become a reality. However, to translate this information to actual clinical improvement, we needed a tool that connects the dots and provides a systems view what is happening in the disease condition.
Dr Brooks-Warburton
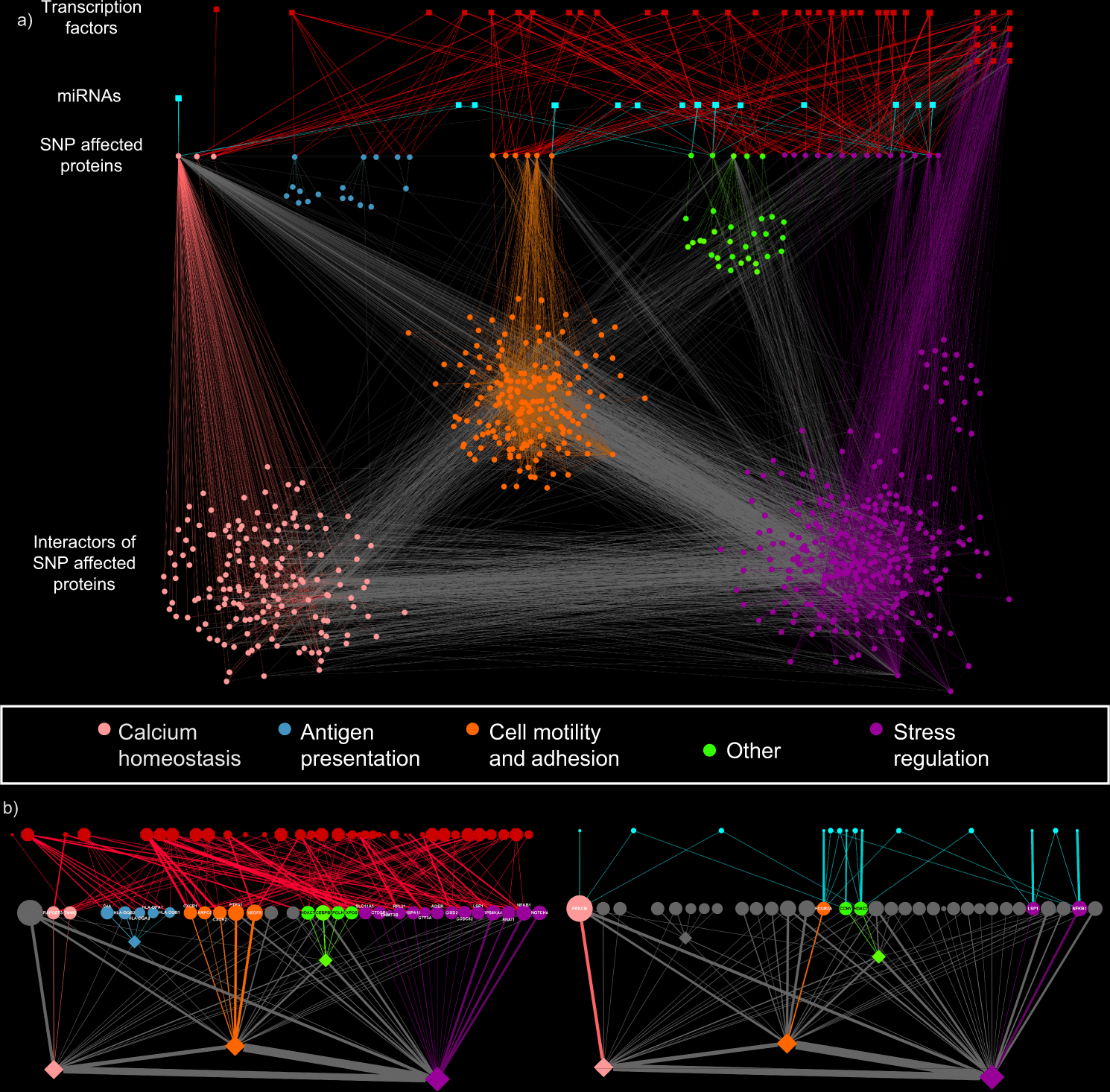
Utilizing SNP profiles of 378 UC patients, the scientists mapped the regulatory impacts of the SNPs to a human signaling network containing protein-protein, miRNA-mRNA, and transcription factor binding interactions.
With unsupervised clustering algorithms, the scientists grouped those patient-explicit networks into four particular groups driven by:
- PRKCB,
- HLA,
- SNAI1/CEBPB/PTPN1, and
- VEGFA/XPO5/POLH hubs.
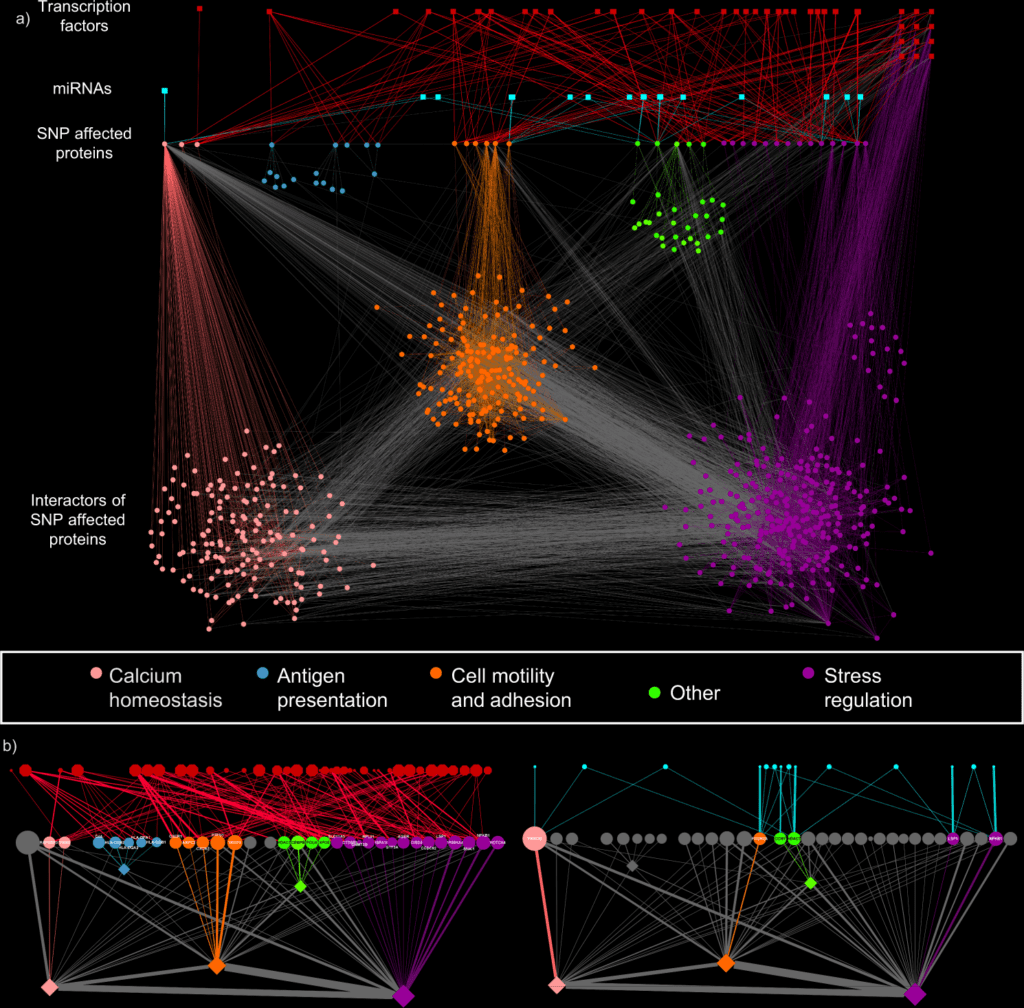
Image Source: A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis.
The pathway analysis identifies calcium homeostasis, wound recuperating, and cell motility as key cycles in UC pathogenesis. Utilizing transcriptomic information from an independent patient cohort, with three complementary validation approaches focussing on the SNP-impacted genes, the patient explicit modules, and affected functions, the researchers confirmed the regulatory impact of non-coding SNPs.
iSNP identified regulatory impacts for disease-related non-coding SNPs, and by predicting the patient-explicit pathogenic cycles, the scientists proposed a systems-level method for stratifying patients.
The power of this approach is that it can find previously unidentified “hidden” connections and proteins in the signalling networks affected by SNPs in the non-coding regions, rather than just those that directly altered genes themselves. It identifies changes in the regulatory mechanisms that previous approaches would miss as they originate in the non-coding regions of the genome.
Dr Dezsö Modos, Quadram Institute
Precision Medicine: Scope and Aim
Precision medicine is a vital clinical aim for the effective treatment of heterogeneous, complex diseases such as inflammatory bowel disease (IBD).
Complex, multilayered, diverse, integrative techniques are required to identify the singular patients’ complex pathogenic pathways. With IBD, the interlinked facets prompting disease are a dysfunctional immune system and response to environmental triggers, including constituents of the intestinal microbiota and dietary factors, in a genetically susceptible host.
Focussing exclusively on genetic susceptibility, genome-wide association studies (GWAS) and subsequent fine-mapping of recognized regions described causal disease-related single nucleotide polymorphisms (SNPs). Notwithstanding, the clinical effect of these SNPs presently is yet to be understood.
A promising way to deal with evaluating the functional role of SNPs, and advise clinical practice, is to examine patient-explicit sets in combination with systems-level approaches.
Identification of the SNPs of Interest
Exome sequencing and protein structural biology have proactively added to the functional annotation of SNPs in protein-coding regions (that alter the amino acid composition and the functionality of the translated proteins) and how they affect diseases such as obesity, IBD, and lung cancer.
Computational workflows focus on such coding SNPs for additional analysis. These methodologies include artificial intelligence strategies (for example, machine learning and deep neural networks) to recognize and quantify deleterious regulatory effects of SNPs utilizing chromatin accessibility and transcription factor binding affinities and high-throughput chromatin interaction studies.
This enables the identification of SNPs of interest. Nonetheless, understanding the function of SNPs in non-coding regions of the DNA stays challenging, basically because numerous disease-causing SNPs are in regions yet to be annotated.
A Need for a Deeper Understanding of Functions of SNPs
In ulcerative colitis (UC), a type of IBD, coding SNPs contain under 10% of the total UC-related SNPs. These coding SNPs are not causally related to impaired intestinal barrier function or inflammation that are trademark pathognomic features of UC.
Comprehension of the phenotypic impacts of SNPs in IBD has involved the study of monogenic IBD in pediatrics that analyzed the deleterious nature of non-coding SNPs, albeit, in adult-onset IBD, these rare individual phenotypic SNPs have not been recognized.
A more extensive and more profound comprehension of the function of SNPs in this complex genetic disease is subsequently required.
The Functional Annotation of SNPs
The scientists proposed that the functional annotation at the molecular and systems-level of the leftover 90% SNPs situated in non-coding regions would expand the utility of the disease-related SNPs.
The proposed gap-filling systems-level analysis is fundamental, as individual SNPs might have subtle phenotypic impacts, yet in combination, they might have a pathological effect.
Integrated analysis of the non-coding SNPs permits the identification of novel pathogenic pathways and possibly patient-explicit disease susceptibility, hence facilitating precision therapy.
For the functional annotation of SNPs in non-coding regions, a key inquiry is whether the SNPs influence gene expression by, for instance, influencing long non-coding RNAs, microRNA-target sites (miRNA-TS), splicing or transcription factor (TF) binding regions (TFBS) in promoter regions and inside the first introns, which has been accounted for in complex diseases, for example, diabetes, schizophrenia, coronary heart disease, and Crohn’s disease.
In this study, the scientists zeroed in on two regulatory impacts as specific examples;
- SNPs happening in transcription factor binding regions and
- In miRNA, target sites can be validated by published documents
Expanding Current Research and Identification of Additional Pathogenic Pathways
To identify the impact of non-coding SNPs, the scientists built on the concepts identified previously to track the cumulative effects of multiple regulatory SNPs as a ‘monogenic’ model.
Utilizing network biology approaches that we have recently taken advantage of to reveal novel and significant proteins in cancer biology, the scientists aimed to additionally understand the pathogenic pathways of UC and to recognize novel and previously hidden disease-related proteins.
These proteins are much of the time undetected or hidden in conventional mutation and expression screens as they, for the most part, act as direct interactors of the proteins impacted by a disease-associated SNP. Utilizing first neighbors gives an ideal trade-off to keep specificity while reconstructing a connected chart.
Similar studies have used the idea of first neighbor proteins in both type 2 diabetes and juvenile idiopathic arthritis. Systems biology approaches have been used with predictive network models that recognize proteins involved in the pathogenesis of IBD in general however, these methodologies can’t represent regulatory and downstream impacts of non-coding SNPs.
Accordingly, by identifying first neighbor proteins in UC, the researchers aimed to extend current research and recognize additional pathogenic pathways of pharmacological use in UC that have been previously undetected or hidden because of a lack of connection with non-coding UC-associated SNPs.
As UC is profoundly heterogeneous, the researchers utilized individual patient information to recognize patient cohorts with similar or different pathogenic pathways of UC.
The Integrative SNP Network Platform or iSNP
Connecting non-coding SNPs to pathways, particularly in a patient-explicit way, is a genuinely necessary yet profoundly challenging approach.
To accomplish this, the scientists created a workflow named the integrative SNP Network Platform (iSNP) by consolidating systems genomics and network biology approach into a scalable system.
They showed its applicability by analyzing a UC-related signaling network and identifying patient clusters with distinct pathomechanisms contributing to UC.
Inside these clusters, they highlighted cluster-explicit key participants, recognizing known and additional proteins and patient-explicit pathways to the disease.
These predicted pathogenic impacts were then validated utilizing transcriptomic information from an independent patient cohort.
Integrating frameworks genomics and network biology information and analysis offers novel biological insights and allows the scalable assessment of patient-specific datasets for precision medicine.
The Endpoint
The scientists designed an integrated systems genomics workflow, named iSNP, to layer patient information from population-wide genomics with network biology and transcriptomics involving UC as a model of a complex genetic disease.
Their objective was to determine the complex genetic background adding to disease pathogenesis for an individual patient. To accomplish this, they initially recognized so far hidden proteins associated with UC pathogenesis, second, they identified key pathogenic pathways for UC, and, thirdly, they determined that patients had similar or different pathological processes in disease development.
This was done with the goal to give insights that could progress personalized medication for patients with UC. This study utilized functional annotation of non-coding SNPs with the integration of transcriptomics and protein interactions at an individual patient level.
For complex diseases, such as cancer, IBD, and other autoimmune diseases, there are always multiple answers. Until now there were only a few tools that could carry out a complex analysis on a patient-specific way and provide various molecular maps of the same disease. With iSNP, it is now a reality to screen large cohorts, understand better the individual pathogenesis stories, and find the best treatment for the right patient.
Prof. Simon Carding from the Quadram Institute and University of East Anglia.
Article Source: Brooks-Warburton, J., Modos, D., Sudhakar, P. et al. A systems genomics approach to uncover patient-specific pathogenic pathways and proteins in ulcerative colitis. Nat Commun 13, 2299 (2022). https://doi.org/10.1038/s41467-022-29998-8 https://quadram.ac.uk/hidden-genetic-connections-for-personalised-medicine-ibd/
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.