Authors from the University of Pennsylvania and others innovated the motif-specific PPI targeting algorithm (moPPIt), a computational framework that integrates BindEvaluator along with Multi-Objective Guided Discrete Flow Matching (MOG-DFM) to design peptides that can selectively bind functional motifs without needing structural priors. It works directly from sequence data instead of requiring known 3D structures, making it powerful for proteins with intrinsically disordered regions (IDRs).
The Need for Precision in Peptide Binder Generative Algorithms
Many proteins, such as disordered proteins (IDPs), fusion oncoproteins, and proteins with active regions, don’t have a stable tertiary structure or deep hydrophobic pockets. This makes it harder for them to be studied under traditional drug discovery methods, which often rely heavily on structure-based docking.
Existing peptide or small molecule design methods often generate binders that stick broadly to a protein but don’t discriminate between functional motifs. This absence of specificity can lead to off-target effects and diminish the therapeutic accuracy of protein binders.
Even after the integration of AI generative models, prior approaches for peptide design were either not guided by specific binding contexts or lacked the ability to balance multiple objectives, such as affinity, specificity, solubility, etc. So, the binders predicted and generated by them were often inactive or non-functional in real biological systems.
To tackle this long-standing bottleneck, Dr. Pranam’s team designed a motif-specific PPI targeting algorithm (moPPIt). A computational framework that bypasses the need for 3D protein structures by working directly with sequenced data. This makes drug discovery inclusive of disordered proteins, which were previously inaccessible.
Detailed Workflow of moPPIt: How it Integrates BindEvaluator and MOG-DFM
Although the idea of working directly on sequenced data is not new, it has been used in Protein language models (pLMs) like SaLT&PepPr, PerPrCLIP, and PepMLM, but none of these frameworks explicitly addressed motif targeting. This meant binders with desirable global properties could be generated, but they couldn’t be directed to distinct functional epitopes on a protein surface. However, moPPIt couples controllable peptide generation with the specific objective of motif targeting using only sequence data.
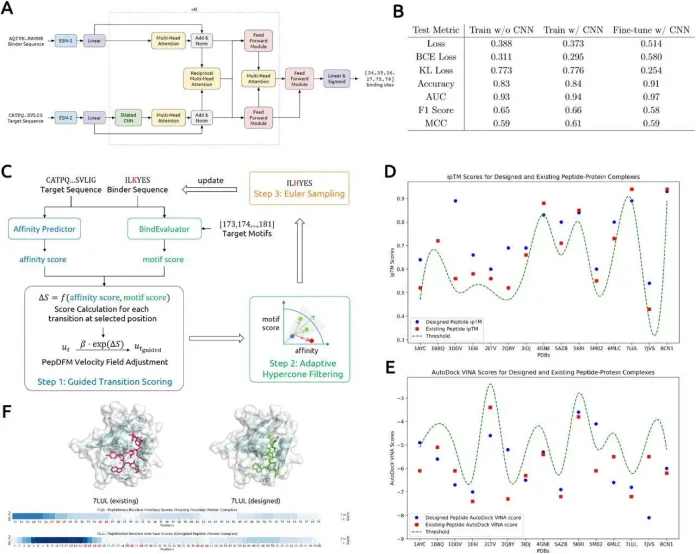
BindEvaluator, a transformer-based model that predicts peptide protein binding sites, is the key predictive engine inside moPPIt. It works by taking two sequences as input: the peptide and the protein. Both sequences are embedded using the pretrained ESM-2-650M protein language model, which encodes biochemical and evolutionary features.
Target embeddings are processed by dilated CNNs to capture residue-level binding features (taking local properties into account) while multi-head attention layers encode long-range sequence dependencies.
At last, reciprocal attention modules fuse binder and target representations, modeling their interactions. These representations are passed through feed-forward layers to predict binding residues on the target.
BinderEvaluator is integrated into Multi-Objective Guided Discrete Flow Matching (MOG-DFM), which directs PepDFM (discrete flow matching peptide generator) toward Pareto-efficient trade-offs. This balances multiple objectives simultaneously.
The signals from bindingEvaluator and Affinity predictors finally transform PepDFM into moPPIt, completing the framework that generates motif-specific peptides with high affinity.
Key Results: InSilico Validation of Peptides Generated by moPPIt
To test moPPIt’s ability to design motif-specific, high-affinity peptide binders, the authors chose the following progressively harder tasks:
- Designing binders for proteins with known binders:
A set of 15 structured proteins from the Protein Data Bank (PDB) was selected as targets. It was ensured that experimentally validated peptides were also not included in the training data.
All designed peptides for the targets met strict ipTM scores (AlphaFold3 interface confidence) within 0.05 of reference complexes, and 12 out of 15 designed peptides showed VINA docking scores >1.0, lower than reference complexes. High-energy residues are consistently localised to target motifs, confirming motif specificity.
- De novo peptide design for epitopes within structured proteins:
Researchers selected a few classes of protein targets to test moPPIt’s ability to design peptide binders for structural proteins: kinases, phosphatases, deubiquitinases, and G-protein-coupled receptors (GPCRs).
Candidate epitopes were selected using APBS electrostatic analysis, which highlights regions of favorable charge for peptide binding.
Designed peptides achieved a mean AutoDock VINA score of -8.2 kcal/mol, indicating strong predicted binding affinities. PeptiDerive RIS analysis also confirmed that binding energies were localised to the target motif sequences. This shows that moPPI’s binders interact specifically with the intended epitopes rather than non-specific regions.
- Peptide binder design for intrinsically disordered regions:
To test moPPIt on intrinsically disordered proteins (IDPs), which are considered undruggable due to their lack of stable structures, the team chose MYC (an oncogenic transcription factor) and EWS::FLI1.
Here, AlphaFold3 predictions indicate that peptides designed by moPPIt showed specific interactions with targeted motifs. Favorable VINA docking scores and high PeptiDerive scores together indicate localized peptide binding, confirming strong, motif-specific engagement.
Conclusion
The study proves that it’s possible to design custom peptide binders directly from protein sequences using moPPIt. By combining both guiding models, BindEvaluator and MOG-DFM, researchers have created a system that achieves both accuracy and strength at once. This opens the door to designing new therapeutic molecules that can block or control protein interactions precisely.
Article Source: Reference Paper | Code Availability: The datasets and codebase to train BindEvaluator and construct moPPIt is freely accessible at HuggingFace alongside an easy-to-use Colab notebook for inference.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.















