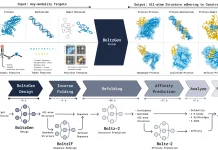
Scientists from MIT and Microsoft Research present CleaveNet, an AI-based pipeline that merges predictive and generative modeling for end-to-end peptide (short protein) design. By validating the system on matrix metalloproteinases (MMPs), the team demonstrates that AI can uncover novel cleavage motifs, engineer selective substrates, and overcome ever-existing challenges in protease promiscuity.
Balancing Efficiency, Selectivity, and the Complexity of Protease Substrate Design
A peptide is typically about 10 amino acids long, with 20 natural amino acids available at each position, which means the total design space is about 1013. That’s 10 trillion possible sequences, which is far too many to test experimentally. This huge space makes it nearly impossible to identify optimal substrates.
Coming to proteases, they are often evolved from a common ancestor, so related enzymes share overlapping substrate preferences. This overlap means that even if a substrate is efficiently cleaved, it may not be selective (multiple proteases could act on it). Designing substrates that are unique to one protease is, therefore, extremely challenging.
The efficiency of a substrate is judged by how strongly it is cleaved by the target protease. Achieving efficiency and selectivity simultaneously is rare because proteases tolerate diverse sequences and hence many substrates end up either efficient but promiscuous or selective but inefficient.
Traditional methods are slow, costly, and too simple to overcome these challenges, considering the complex and context-dependent biochemistry of substrate design.
How CleaveNet Tackle’s Protease Design Challenges
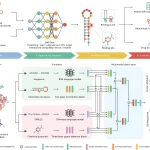
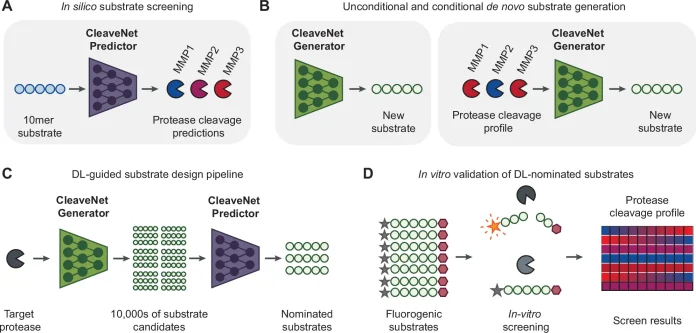
CleaveNet is an AI-based system designed specifically for protease substrate design. It combines two components. A predictor, which is a deep learning model that predicts how efficiently a peptide will be cleaved by different proteases, and a generator, a generative model that creates new peptide sequences, either freely (unconditional) or guided by desired cleavage preferences (conditional).
Together, they allow researchers to nominate, evaluate, and design substrates entirely in silico.
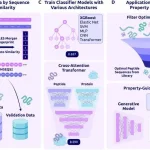
Understanding Architectures of CleaveNet Predictor and Generator
The authors tested two different architectures for CleaveNet’s predictor model, LSTM and Transformer. Out of which the transformer was chosen for the best performance and scalability. The predictor was trained on a dataset having ~18,500 synthetic peptides from mRNA display libraries and screened against 18 matrix metalloproteinases (MMPs).
Output:
- Continuous Z scores representing cleavage strength
- An optional binary classification (cleaved vs not cleaved) based on thresholds
- Uncertainty estimates via deep ensembles (5 predictors).
Predictor showed strong correlation with experimental cleavage data (Pearson’s r of 0.8 for MMP13) and robustness across different test sets, including independent fluorescence assays. It held up to its reliability with a ROC-AUC of up to 0.98 at higher thresholds.
Generators architecture, on the other hand, is an Autoregressive transformer trained on the same mRNA display dataset. The sequences generated matched amino acid distributions and biophysical properties to those of read peptides, revealed new preferences, and showed k-mer diversity similar to experimental libraries.
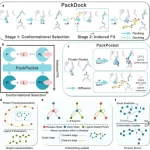
Experimental Validation of CleaveNet
95 fluorogenic substrates designed by Cleavenet were tested against recombinant MMPs.
As a result, all CleaveNet-designed substrates were cleavable by MMP13, out of which some conditionally designed substrates were uniquely cleaved, showing true selectivity.
Limitations and Future Directions
CleaveNet is trained on short synthetic peptides generated using mRNA display libraries. This means it only understands cleavage rules for short substrates. Real protease systems involve longer sequences that the model hasn’t really seen.
Another limitation is that the model was tested on MMPs. Whether CleaveNet performs equally well on other protease families remains an open question.
Future versions of CleavNet could work with longer sequences or go beyond proteases by training on a wider range of datasets. CleaveNet. could also combine sequence-based learning, which would help understand the 3D context of protease substrate interactions, improving accuracy.
Authors also plan to make CleaveNet usable for labs that don’t have access to expensive display technologies. This could speed up substrate design, especially where resources are limited.
Final Takeaways
CleaveNet is a deep learning pipeline that combines prediction and generation to design protease substrates. It has been trained on a large experimental dataset, and can accurately predict cleavage efficiencies, generate novel plausible sequences, and validate them experimentally. By understanding new rules of efficiency and selectivity, CleaveNet addresses challenges of proteases’ promiscuity and opens the door to scalable, tunable, and accurate substrate design.
Article Source: Reference Paper | Reference Article |Availability: GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.