A project coordinated by a group of researchers from the Eichman lab and the Evolutionary Studies Initiative on the research of chemical compounds produced by bacteria was just published in mBio.
Image Source: Resistance-Guided Mining of Bacterial Genotoxins Defines a Family of DNA Glycosylases.
The group was particularly interested in a class of compounds known as natural products. These substances are created by an organism for a specific function, but they are frequently valuable because the undiscovered beneficial therapeutic chemicals can be employed as antibiotics, anticancer agents, or other treatments. As new drug-resistant disease strains emerge, the need of developing new weapons to combat resistant diseases grows.
Bacteria produce an abundance of secondary metabolites, which are essential for their existence and frequently have medicinal benefits. Streptomyces produces roughly 70% of all commercially effective antibiotics and 35% of all known microbial natural products, with numerous FDA-approved antitumor medicines used as first-line cancer therapy.
Secondary metabolites are frequently exploited as toxins in ecological interactions with other organisms, and they can target a variety of important cellular activities. Natural products that cause DNA damage (genotoxins) generate covalent or noncovalent DNA adducts that can obstruct replication and transcription, compromising genomic integrity through mutagenesis or cell death. As a result, genotoxins are particularly effective anticancer medicines, as demonstrated by doxorubicin, bleomycin, mitomycin C, and duocarmycin analogs, among other clinically relevant medications.
In several organisms, resistance genome mining has emerged as a crucial bioinformatically driven pipeline for discovering novel natural products and gene clusters. The substantially reduced candidate pool as a consequence of the targeted discovery of gene clusters containing a resistance gene is a key benefit of resistance genome mining.
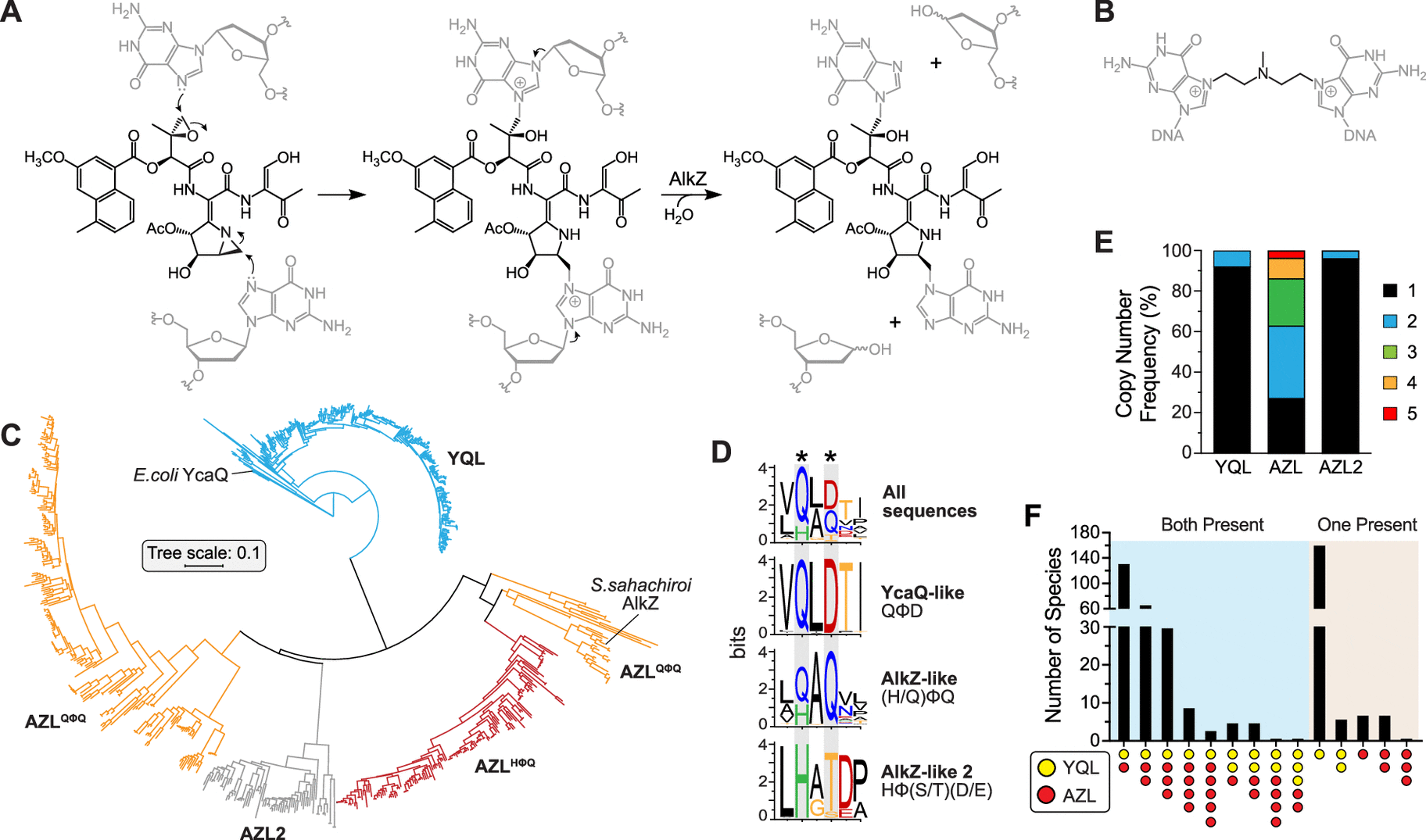
Genome mining is a powerful tool for discovering gene clusters and the natural compounds they produce. Genome mining has traditionally been used to target a specific mechanism that produces a natural product. However, because numerous systems can produce comparable natural products, the team’s approach focuses on processes that provide bacteria self-resistance to a specific molecule. They were particularly interested in looking for gene clusters that produce genotoxic natural products, or those that can bind to DNA chemically.
The researchers focused on two DNA glycosylases, which are bacterial DNA repair enzymes.
The AlkZ gene is present in Streptomyces sahachiroi, while the YcaQ gene is found in Escherichia coli. These enzymes, as well as others related to them, can repair DNA damage induced by genotoxins. AlkZ and YcaQ, on the other hand, have substantially diverse genomic contexts and roles.
AlkZ-like (AZL) enzymes tend to localize within biosynthetic gene clusters (BCGs) and according to Bradley, “AZL proteins seem to be tailored for self-resistance to specific natural products.”
However, YcaQ-like (YQL) enzymes have not been found in clusters and according to Katie Wahl, another co-first author of the study, “YQL proteins might be general caretakers to remove a variety of DNA damage.”
“In the short term, we hope that the approximately 70 uncharacterized gene clusters we identified in our study can be screened for the targeted discovery of novel or useful genotoxic natural products,” said Bradley.
“In the long term, we are hopeful that our self-resistance-guided genome mining framework will be applied in the scientific community for the targeted discovery of natural products of therapeutic benefit. We also envision the discovery of new DNA repair mechanisms through this approach,” added Wahl.
The discovery of new analogs or types of DNA alkylating/damaging secondary metabolites from these orphan clusters could be a crucial step in the development of new anticancer or antibacterial medicines. This classification of YQL/AZL proteins in Streptomyces is an important first step toward understanding their evolutionary relationship to one another and to different types of gene clusters, and it shows that targeted resistance genome mining is a viable method for discovering novel genotoxins and resistance mechanisms from uncharacterized gene clusters.
Story Source: Bradley, N. P., Wahl, K. L., Steenwyk, J. L., Rokas, A., Eichman, B. F., Simmons, L. A., & Bush, K. (n.d.). Resistance-Guided Mining of Bacterial Genotoxins Defines a Family of DNA Glycosylases. MBio, 0(0), e03297-21. https://doi.org/10.1128/mbio.03297-21 https://www.vanderbilt.edu/evolution/vanderbilt-researchers-use-new-method-to-target-potentially-undiscovered-beneficial-therapeutic-chemicals/
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.