PROTLLM is a versatile crossmodal large language model (LLM) proposed by Beijing Institute of Technology researchers designed for protein-centric and protein-language tasks. It can handle complicated inputs with natural language text mixed with an arbitrary number of proteins thanks to its dynamic protein mounting technique. By creating a specialized protein vocabulary, the proteins-word language modeling approach teaches PROTLLM to predict proteins from a large candidate pool. For pre-training, the extensive interleaved protein-text dataset, known as InterPT, is built using both structured and unstructured data sources. In protein-centric tasks, PROTLLM outperforms baselines with specialized knowledge of proteins, and in protein-language tasks, it fosters zero-shot and in-context learning abilities.
Introduction
Research in the biosciences and the unraveling of life’s secrets depend on an understanding of proteins. Artificial intelligence systems that utilize protein-centric applications such as function prediction and protein folding prediction are made possible by deep learning techniques and neural network models. For protein representation learning, LLM is an effective method that makes tasks like function description generation possible. It learns unsupervised protein representations on big protein sequences using masked language modeling and large-scale pre-training. In recent studies, zero-shot text-to-protein retrieval has been accomplished through the extension of protein models to protein language scenarios. An instruction dataset for the biomolecular domain is presented by Fang et al. (2023).
Protein representation techniques have shown potential in certain applications; however, protein-centric and protein-language tasks remain difficult to model because of their unique architectures, dependence on cross-modal supervision from annotated protein-text pairs, and a variable sequence number that causes pre-training inefficiencies and computational uncertainty. Protein representation techniques are still being investigated for their potential in language and protein-centric tasks despite these obstacles.
Key Features of the Study
- PROTLLM is proposed as a versatile crossmodal LLM for protein-centric and protein-language tasks, capable of processing complex inputs and outputs.
- A large-scale pre-training dataset called InterPT is introduced, integrating both structured and unstructured inputs from scientific research.
- Outperforming protein-specialized baselines and exhibiting zero-shot and in-context learning capabilities, PROTLLM produces better performance in protein-centric tasks.
Understanding PROTLLM
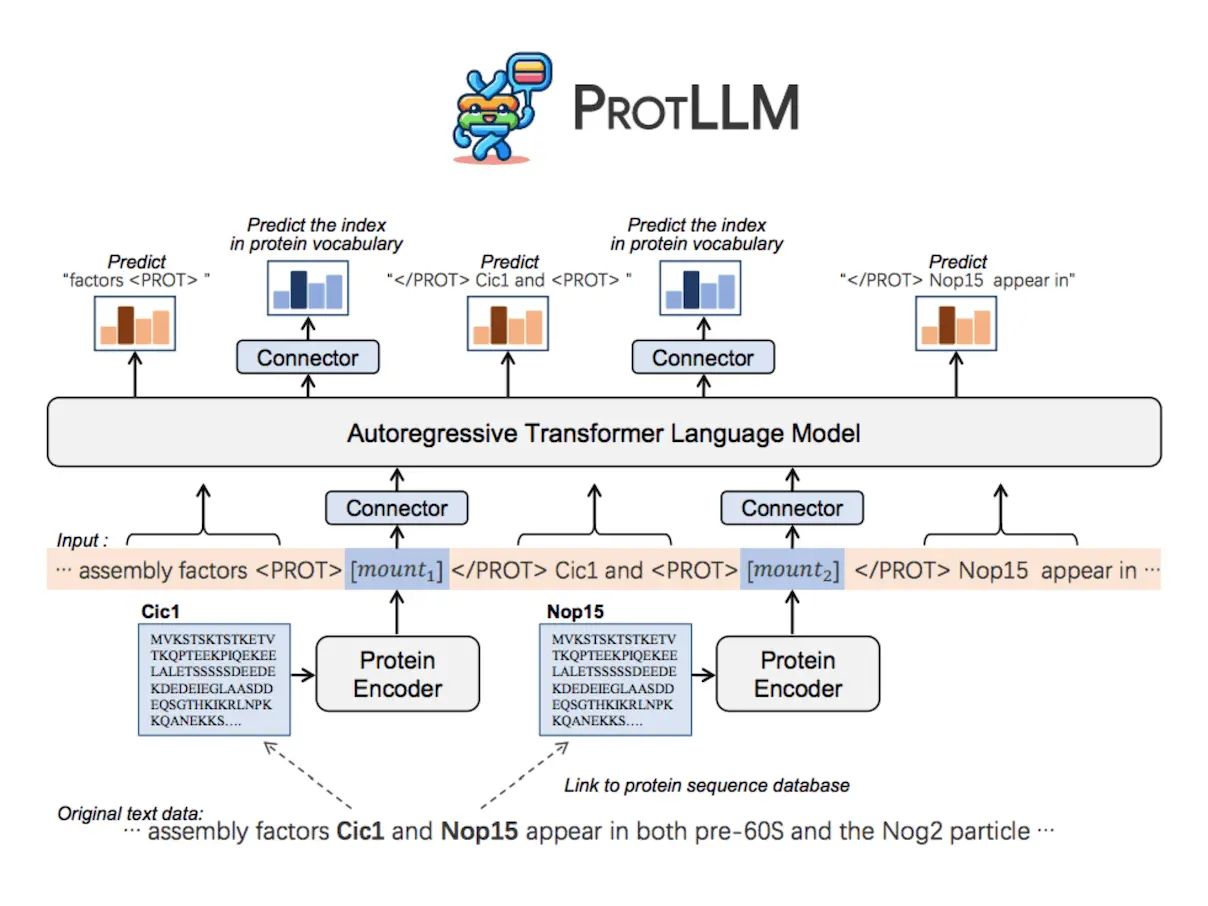
PROTLLM is a flexible LLM model that is intended for protein-centric and protein-language applications. Because of its ability to handle intricately interwoven protein-text inputs and outputs, the model can manage a variety of downstream jobs without the need to redesign task-specific architecture. Text containing an infinite number of proteins can be processed with ease with the help of the dynamic protein mounting technique. In order to guarantee interleaved protein-text outputs, PROTLLM additionally uses protein-as-word language modeling, which autoregressively predicts words and proteins from their corresponding vocabularies.
InterPT: Interleaving Protein-Text Data
PROTLLM is able to use important information from scientific papers for pre-training thanks to InterPT, a large-scale protein-text collection that is derived from both structured and unstructured sources, including coupled protein annotation data. Three different types of data sources are included in this dataset: protein-annotation pairings, multi-protein scientific articles, and protein instruction following data.
Large Language Models
Recent advances in natural language processing (NLP) have demonstrated that these models are capable of tasks beyond text-based tasks and, thus, beyond the domain expertise of humans. These models have been applied to complex decision-making, drug development, and mathematical problem-solving. A crucial method for lining up pre-trained encoders with LLMs so they can comprehend multimodal inputs is instruction tuning. For applications including retrieval, captioning, and molecular property prediction in scientific research, specialized molecular LLMs have been developed. However, the lack of complete datasets and ineffective architectures to model protein-language sequences continues to impede progress in understanding proteins with LLMs.
Protein Representation Learning
Understanding proteins through protein representation learning and protein language models (PLMs) has advanced significantly in protein modeling. In order to acquire coarse-grained representations at the amino acid level or fine-grained representations at the atom level, these models train protein sequence encoders on large amounts of protein sequence data. Nevertheless, text data pertaining to proteins is frequently disregarded, although possessing important supervision signals that are required to comprehend proteins. With multimodal pre-training on protein-text pairs, OntoProtein incorporates textual information into the protein encoder to improve protein understanding with text supervision. It does this by utilizing knowledge graphs and ProtST. By providing an extensive instruction dataset for biomolecules, Mol-Instruction improves LLM performance even further. Using a structured knowledge graph as a sample, InstructProtein enhances the quality of instruction datasets. However, these approaches have limitations, such as their direct incorporation of protein sequences into LLMs as text, leading to suboptimal protein modeling.
PROTLLM Architecture
A protein encoder, cross-modal connectors to comprehend protein sequences, and an LLM for natural language modeling are all combined to create PROTLLM, a natural language model. ProtST, the protein encoder, adds a two-layer MLP projection head while maintaining the ESM-2 backbone architecture. ProtST uses contrastive learning to learn protein representations that are aligned with text by analyzing large-scale protein-text pairs. The LLM and the protein encoder are connected via cross-modal connectors, which allows PROTLLM to receive multimodal inputs. Protein representation space output vectors are transformed into LLM representation space by the input-layer connector and back into protein representation space by the output-layer connector. The output-layer connector also serves as a prediction head, enabling protein retrieval and multichoice protein answering tasks without requiring the LLM to generate complicated protein names.
In-Context Learning in PROTLLM
The work shows that a human protein-protein interaction (PPI) model can learn in context for tasks that focus on proteins. As the number of demonstration cases increases, the model constantly attains higher accuracy, indicating its efficacy. It does not, however, learn in context with 2, 6, and 12 examples and difficulties with scientific publications involving several proteins. The in-context learning of the model can be useful for specific tasks in the absence of labeled data.
Conclusion
Using a natural language interface, PROTLLM is an adaptable LLM that manages intricately interwoven protein-text data, bringing together a diverse range of protein functions. In order to promote learning from a variety of data sources, it builds the extensive protein-language pre-training dataset InterPT. PROTLLM’s competitive performance versus specialized baselines is demonstrated through extensive experiments, opening up new avenues for protein-language applications and showcasing the platform’s versatility.
Article source: Reference Paper | PROTLLM: Project | GitHub
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.