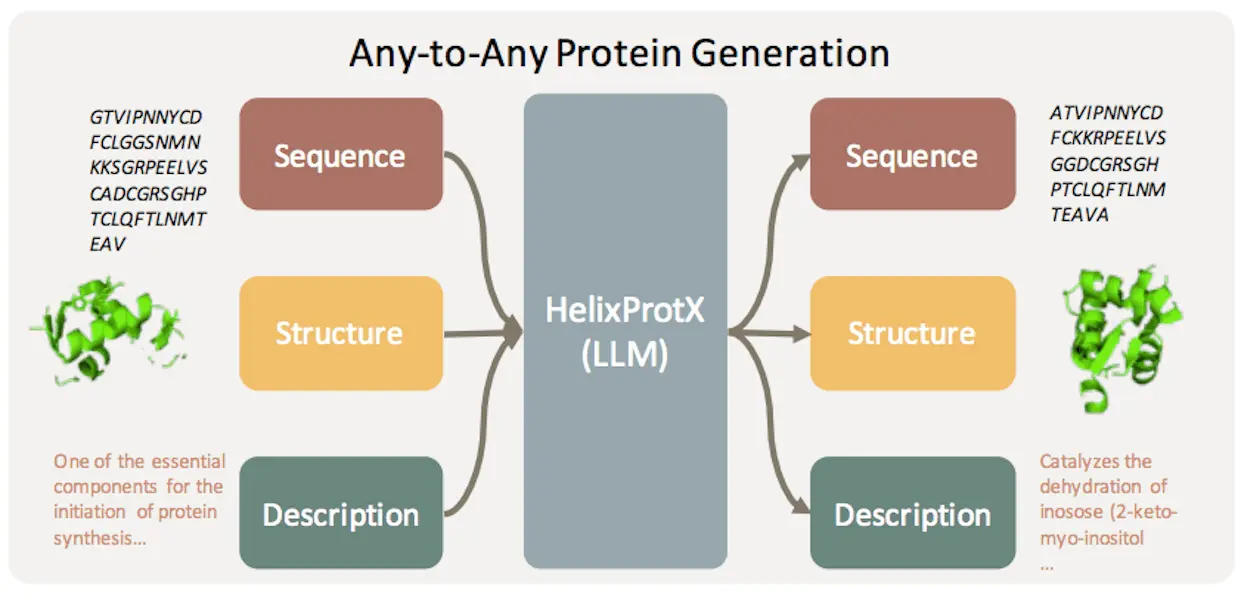
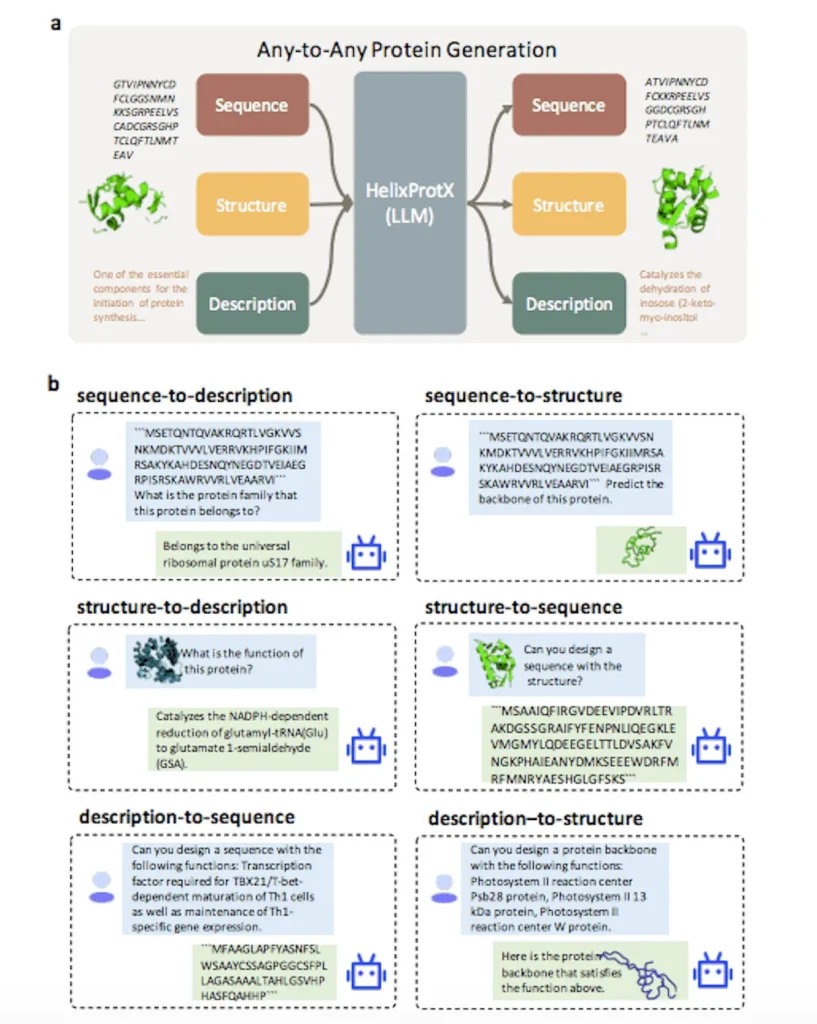
In an effort to provide a comprehensive solution for protein research, researchers present HelixProtX, a system built around the large multimodal model that facilitates the construction of any-to-any protein modality. It enables the conversion of any input protein modality into any desired protein modality, in contrast to current approaches. The experimental results confirm HelixProtX’s superior abilities in performing important tasks, including building protein sequences and structures from textual descriptions and producing functional descriptions from amino acid sequences. According to preliminary results, HelixProtX routinely outperforms current state-of-the-art models in terms of accuracy across a variety of protein-related tasks. HelixProtX promises to speed up scientific research by introducing multimodal large models into protein research, creating fresh perspectives on protein biology.
Introduction
Proteins are basic building blocks of biological systems and can be represented in a number of ways, such as text descriptions, structures, and sequences. Although deep learning and scientific large language models (LLMs) for protein research have made significant strides, present approaches mostly concentrate on narrowly specific objectives, frequently predicting one protein modality from another. These methods limit the creation and comprehension of multimodal protein data. Large multimodal models, on the other hand, have shown promise in producing any-to-any content, such as text, photos, and videos, enhancing user interactions in a variety of contexts. The use of these multimodal model technologies in protein research holds great potential since it could revolutionize how proteins are investigated.
Methods to Study Proteins
Proteins can be studied using a variety of methods, such as textual descriptions, three-dimensional structures, and amino acid sequences, each of which provides a distinct perspective on how to comprehend proteins: (1) The fundamental structure of a protein, known as its amino acid sequence, is responsible for encoding genetic information. Sequence-based deep learning models, like those described, are commonly utilized to examine the relationships between residues. (2) The folded state, or three-dimensional structural conformation, has a significant impact on a protein’s functional activity. A number of sophisticated structural encoders have been studied to clarify the intricate spatial connections between residues or atoms within a protein, including the Geometric Vector Perceptron (GVP), Protein Message-Passing Neural Network (ProteinMPNN), and Invariant Point Attention (IPA). (3) The narrative viewpoint on protein functions, as reported in the scientific literature, is provided by the textual description. This textual modality offers insightful information that broadens the comprehension of proteins’ various functions and traits.
Deep Learning in Protein Generation
The primary focus of deep learning-based protein research is on singular-task approaches to predict one protein modality from another. With the ability to predict three-dimensional protein structural conformation from amino acid sequences, AlphaFold II and RoseTTAfold have greatly advanced protein structure prediction. With the goal of producing sequences that can fold into particular three-dimensional structures and producing protein sequences or structures based on a given tag or description, AI-driven protein design has attracted a lot of scientific interest. However, existing models usually focus on specific protein-related activities, which makes the research process more difficult and burdensome. Such a comprehensive approach that tackles several protein-related issues at once will greatly simplify matters and expedite research. This would facilitate the management of various input and output formats, the understanding of many models, and the reconciliation of possibly contradictory findings across models.
Understanding HelixProtX
HelixProtX is a complete protein research system that integrates descriptions, structures, and sequencing of proteins. Because it is based on a large multimodal model, any-to-any protein creation is possible. The main objective is to modernize protein research procedures and make them more thorough and effective. HelixProtX seamlessly integrates sequences, structures, and textual descriptions to enable the production of several protein modalities from any input. The system generates accurate and comprehensive responses across several protein modalities by analyzing the semantics and context of each question.
The protein-related task tool HelixProtX has shown promise in a wide range of applications. In most tasks, it performed better than the current standards by utilizing the sophisticated features of large multimodal models. The approach showed remarkable potential in text-guided protein design applications and proved resilient across proteins of different lengths and families. The proteins that were developed exhibited coherence and logic by closely matching reference proteins. HelixProtX has the potential to expedite scientific discovery in protein biology by offering new insights into the realm of protein biology through the use of substantial multimodal models for any-to-any protein creation.
Conclusion
HelixProtX is an integrated large multimodal model system that has demonstrated superiority over baseline approaches in protein description prediction and sequence design tasks by successful inter-mapping of several protein modalities. The substantial promise of large multimodal models in the biological sciences is confirmed by this work, opening up fresh perspectives on protein biology and hastening scientific advancements. Subsequent investigations should concentrate on enhancing the prediction and design of structures and expanding the range of applications to encompass supplementary life science fields such as RNA and tiny molecules. HelixProtX can better represent the intricacies of protein spatial structures and improve its performance in protein-related activities by investigating more complicated network topologies and sharpening optimization goals.
Article Source: Reference Paper | The source code and inference code of HelixProtX are freely available on GitHub.
Important Note: ChemRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.