A team of researchers from the University of Washington has made significant strides in protein design by addressing long-standing challenges in creating beta-barrel structures. Using advanced deep learning tools, including RoseTTAFold-based design techniques, the team successfully developed transmembrane nanopores with conductances ranging from 200 to 500 pS. This innovative approach combines the precision of parametric generation with the power of deep learning, offering unprecedented control over global protein shapes and expanding the potential for novel protein engineering.
Beta-barrel structures, known for their utility in creating fluorescent proteins and transmembrane nanopores, have traditionally been difficult to design due to deviations from ideal geometries. While previous methods relied on 2D structural blueprints, they provided only indirect control over shape and required significant expertise. The University of Washington team overcame these limitations by utilizing tools like RFjoint inpainting and RFdiffusion, which exploit the intricate sequence-structure correlations inherent in deep learning models.
This breakthrough enables the design of beta barrels with minimal deviation from idealized forms, marking a transformative step in synthetic biology. By seamlessly integrating parametric representations with deep learning, the researchers have unlocked new possibilities for creating proteins with complex global shapes, paving the way for innovations in bioengineering and therapeutic development.
Bridging Classical Design and Deep Learning for Beta-Barrel Protein Design
Many different architectures and functionalities have been produced by using the Crick parametrization of coiled coils to create protein backbones. Even when backbones are loosened to enhance hydrogen bonding by Rosetta-based structural relaxation, this method does not provide designs that fold when created in the lab. As an alternative, Rosetta “blueprints” have been used to generate beta barrels with novel structures and capabilities. With this method, the overall form can only be indirectly controlled by explicitly specifying blueprint elements such as barrel shear, strand lengths, strand numbers, backbone hydrogen bonding arrangement, and irregularity placement. Even with human knowledge, the investigation of beta-barrel structure space is limited by this complexity.
Compared to traditional energy-based techniques like Rosetta, protein folding performance has been greatly enhanced using RFjoint inpainting and other deep-learning techniques. These neural network-based techniques are able to incorporate abnormalities into intended beta barrels by implicitly parameterizing protein folding regulations. However, deep-learning techniques have not been able to provide exact parameter-based control on global fold geometry. This emphasizes the necessity of fusing the advantages of deep learning (greater computational and experimental success rates, more design complexity, and ease of use) with the advantages of classical design (accurate control of structure parameters).
Leveraging Parametric Design and Deep Learning for Beta-Barrel Refinement
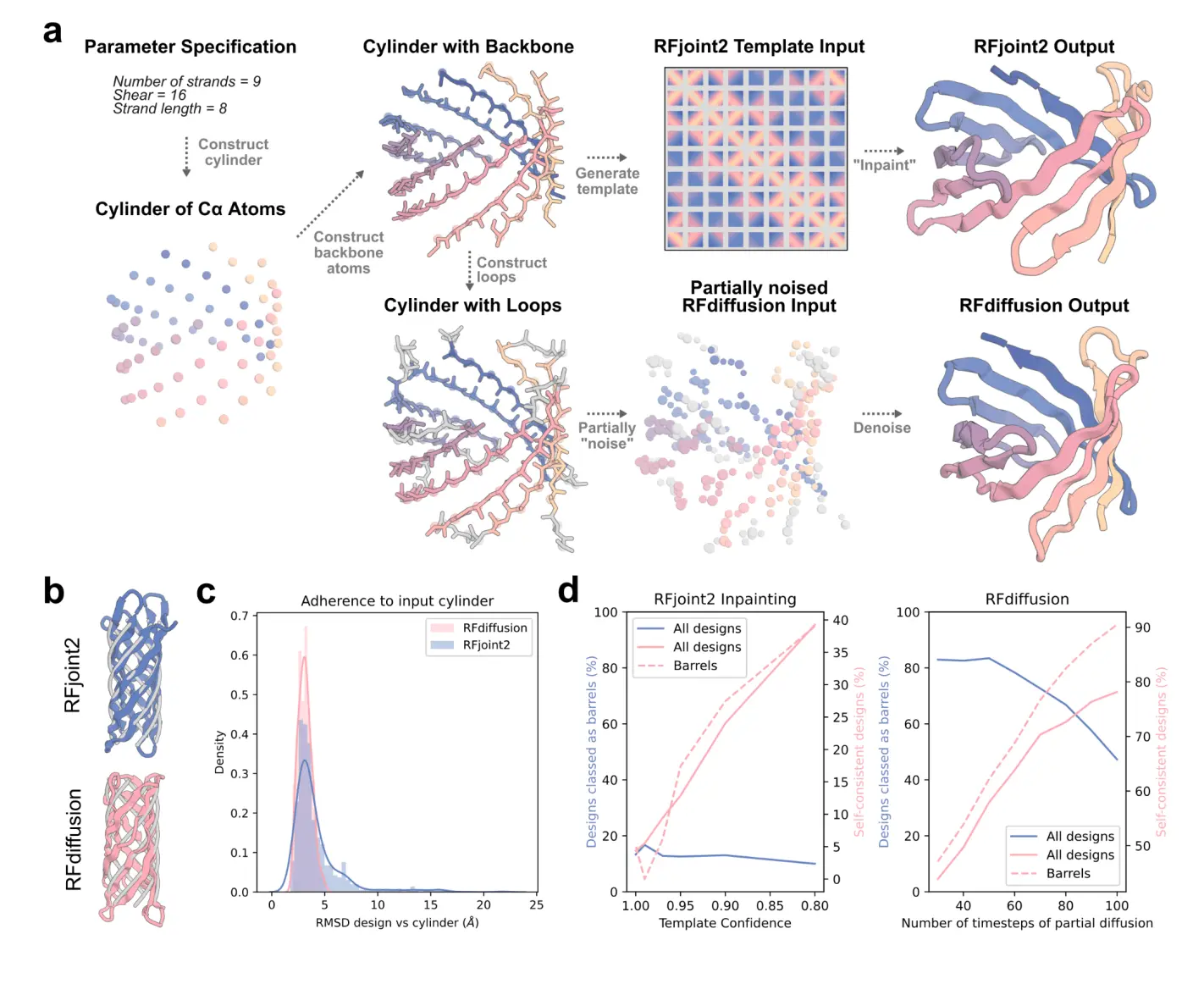
Researchers in this study reasoned that by first using parametric design to generate backbones and then using deep learning techniques to refine them, beta-barrel design based on global parameterization of barrel size and shape could be made possible by the deep understanding of sequence-structure relationships implicit in deep learning based design approaches and their capacity to be guided by partial structural information. To create beta barrels using global parameterizations of beta-barrel shape, researchers looked into both inpainting and diffusion techniques.
Researchers first investigated the possibility of designing beta barrels using faulty templates as input structures using RFjoint2, an enhanced variant of RoseTTAFold-based RFjoint inpainting. RFjoint2 is a network trained to address the motif-scaffolding problem, where a protein substructure is provided and the scaffold is generated. The network uses 2D inter-residue pairwise distances and pairwise dihedral angles to generate large scaffolds around a small input motif. The underlying RoseTTAFold model can take input homologous structures (templates) and a one-dimensional “confidence” feature, indicating how similar the template is to the query sequence. This notion of an “imperfect” template input is maintained in RFjoint2, refining imperfect input structures and confidence values.
The research produced C3 atom cylinders using input templates that varied in strand number (n), shear number (S), register shift (r), and strand length (l). Because of flaws and differences in beta-strand lengths, these sites are not ideal but roughly correspond to the beta-barrel fold. It was predicted that RFjoint2 would add the features and flaws required for correct folding while preserving the intended parameters. RFjoint2 completely inpainted loops linking strands and terminal extensions that were not included in the input template. The N and C backbone atoms needed for the RoseTTAFold template input and initialized 3D frames were added to the Ca cylinders using BBQ (Backbone Building from Quadrilaterals). RFjoint2 should be able to refine these positions even though they are not physically feasible. No sequence input was given to it to make use of RFjoint2’s ability to condition on structural inputs in the absence of sequence.
Conclusion
The study’s main idea is that non-realizable inputs can be refined into designable outputs using deep-learning design techniques like RFdiffusion and RFjoint2. Beta barrels, which have historically been difficult to build computationally, were used to evaluate these approaches. The main idea is confirmed by the experimental characterization of folding, the presence of backbone anomalies in the outputs, and the x-ray crystal structure. By implicitly learning the principles necessary for beta-barrel folding, both models achieved previously unheard-of speeds and success rates (often greater than 50% in silico success) over a wide range of barrel characteristics. The RoseTTAFold structure prediction network serves as the foundation for the initial training of the RFjoint2 and RFdiffusion models, utilizing the representations acquired during training without requiring a lot of computation. By using pre-existing protein networks without retraining, these models are used in a way that is unrelated to the tasks on which they were trained. Like huge language models used for domain-specific fine-tuned models, protein structure prediction models are becoming the foundation models for protein design.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.