The graceful, rhythmic beating of slender cellular appendages called flagella allows sperm cells and microorganisms like algae to swim. But how these waving motions spontaneously self-organize has confounded science for decades. Now, researchers at the University of Bristol propose flagella beating dynamics can emerge from basic physical interactions between motors and elastic elements, described by a simple “reaction-diffusion” theory like those underlying spirals in other natural systems. Their model quantitatively reproduces beating patterns of evolutionarily distant flagella, suggesting common physicochemical origins across species.
The Puzzle of Self-Organized Flagella Dynamics
Flagella are hair-like cellular protrusions filled with an internal “motor” structure termed the axoneme. The axoneme comprises arrays of protein filaments called microtubules that can slide past each other when molecular motors called dyneins pull them together. Somehow, coordinating the activity of thousands of dyneins transforms these local shearing motions into propagating bends along the flagellum.
But figuring out how flagella spontaneously adopt rhythmic beating has challenged science for a long. Proposed ideas like motors sensing local curvature lack definitive proof. Modeling the intricate mechanics and feedback mathematically has been enormously difficult.
This study puts forth that a simple “reaction-diffusion” theory akin to those describing spirals formed by chemical interactions can capture the emergence and diversity of flagellar dynamics across species. Using simulations, they show quantitative agreement with experimental measurements, potentially explaining the self-organization of beating from fundamental physical principles.
Reaction-Diffusion Systems Pervade Nature’s Patterns
Reaction-diffusion theories mathematically depict how local activation and inhibition reactions can spread spatially via diffusion, generating emergent heterogeneity even from identical starting conditions.
These models efficiently describe diverse phenomena like animal coat patterns, oscillating chemical reactions, and ecological predator-prey cycles by specifying just a few reactive chemical species and their diffusion capacities. By tuning reaction and diffusion terms, varied patterns manifest like spots, stripes, and spirals, reflecting those actually observed in nature.
The crucial insight enabling the application of these simple but powerful theories to flagella beating is that just like reactive chemicals, the mechanical forces generated can also diffuse via elastic couplings to nearby regions. This conceptualizes flagella as reaction-diffusion systems in their own right.
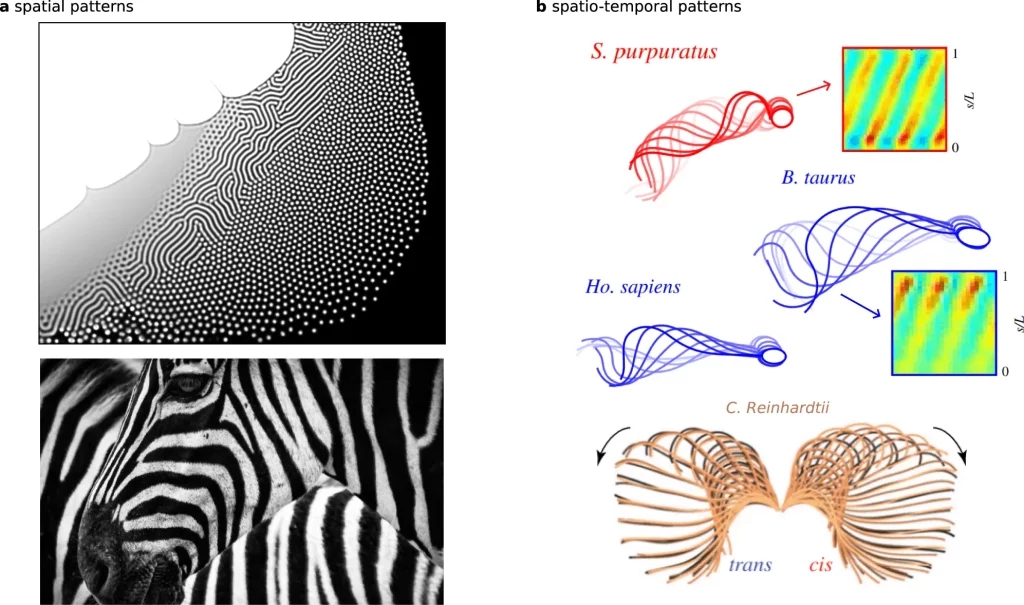
Image Source: https://doi.org/10.1038/s41467-023-40338-2
Deriving a Minimal Theory for Flagella Beating
The researchers leveraged a robust microscopic model of motor proteins laboriously pulling flagellar microtubules via transient attachments. This physiologically faithful model incorporated flexural resistance from flagellar elasticity and fluid drag forces that enable propulsion.
Through mathematical examination of the relative contributions and scales of the internal protein forces and external fluid forces, they derived a greatly simplified “reaction-diffusion” system. This describes a balance of just four key factors:
- Molecular motor activity generating shearing forces
- Internal resistance slowing motor motions
- Elasticity transmitting shearing as bending to nearby regions
- Bending resistance to spatial deformation
This minimal framework mathematically formalizes how local oscillations can propagate as waves – a question posed but unresolved for decades. Intriguingly, varying just three microscopic motor parameters generates diverse beating gaits.
Simulations Reproduce Diverse Flagellar Waveforms
The researchers computationally implemented their derived reaction-diffusion model with these four components. Numerical simulations readily replicated symmetric beating waves akin to algal flagella and asymmetric gaits resembling sperm cells at different microscopic motor settings.
Notably, characteristic waveforms the theory produces quantitatively match measurements from beating flagella of evolutionarily distant unicellular species – Chlamydomonas reinhardtii algae and bull sperm cells. These species have radically different anatomies and flagella lengths, suggesting common general principles independent of organismal details.
The success of capturing different organisms’ beating patterns with the same model contrasts previous indications of fundamentally distinct regulatory mechanisms. This further strengthens the case for scale-bridging theories connecting molecular activities to higher-level biological dynamics.
Additional Factors Modulate Emergence and Beating
The study focuses on the spontaneous generation and propagation of oscillations through reaction kinetics and diffusion alone. However, adding terms for other documented factors like basal stiffness or 3D geometry can quantitatively fine-tune matching experimental shapes.
Intriguingly, having derived their theory from universal conservation principles, the researchers further show it accurately predicts trajectories of freely swimming cells over multiple beating cycles. This verifies that slender moving filaments can self-organize dynamics solely from internal jostling without requiring external fluid feedback.
These findings also provide templates for generating controlled motion in synthetic biological microswimmers, which is potentially helpful for non-invasive therapeutics delivery and diagnostics inside patients.
A Common Basis for Microscopic Waving Structures?
This research constitutes possibly the first precise mathematical demonstration that reaction-diffusion principles central to patterning static systems also directly extend to animate biological structures.
By substantiating that only simple reactive building blocks prove sufficient to generate propagating flagellar waves spontaneously, the study opens avenues to broadly understand dynamic order emerging from collective particle interactions across scales.
Article Source: Reference Paper | An implementation of the reaction-diffusion model is available on GitHub
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.