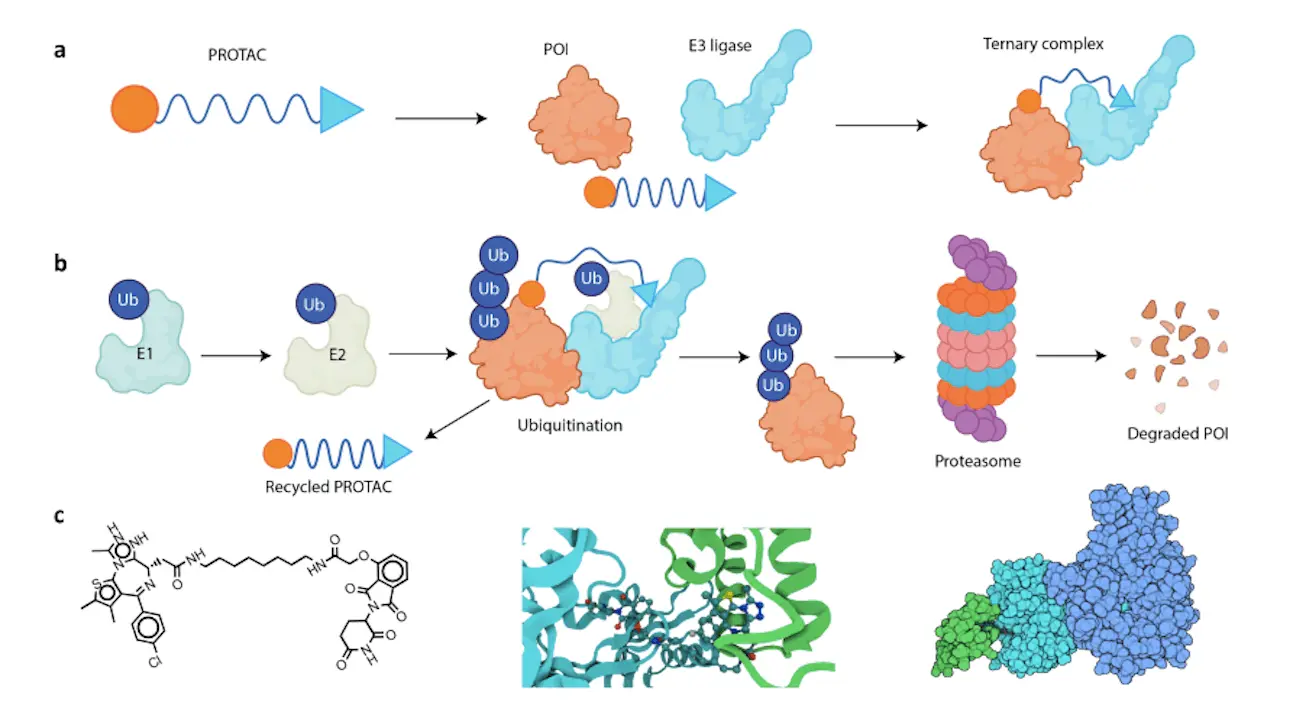
As the fight against disease has changed, so have therapeutic strategies. One such new approach is Targeted Protein Degradation (TPD), which attempts to exploit the cell’s natural degradation machinery to get rid of unnecessary proteins. The cornerstone of TPD is PROTACs (Proteolysis-Targeting Chimeras); these molecules are very complex and act like Trojan horses that bring together a target protein with an E3 ligase, which is a cellular death sentence for proteins. Nevertheless, designing effective PROTACs has been challenging and time-consuming in the past. Consequently, Machine Learning (ML) is now changing this landscape of PROTAC development.
The Power of PROTACs: Beyond Inhibition
Unlike conventional drugs that inhibit protein activity, PROTACs subvert the endogenous protein disposal system within cells. Acting as molecular links between a particular protein (the target) and an E3 ligase; when attached, the E3 ligase tags the target protein for proteasomal degradation, leading to removal from the cell.
This method has various advantages:
Drugging “Undruggable” Proteins: It is difficult to target some proteins with normal inhibitors because they lack well-defined active sites. PROTACs can get around this problem by using the degradation pathway.
Increased Efficacy: Since a single PROTAC molecule may lead to the degradation of several target proteins, they may be more potent than traditional inhibitors.
Prolonged Actions: PROTACs remove the protein entirely, and their therapeutic effects last long.
The Challenge: Designing the Perfect PROTAC
As promising as they are, however, developing effective PROTACs is a challenge. These molecules have three key components:
- Warhead: This section identifies and binds itself to the target protein.
- Linker: An elastic part that joins the warhead with the E3 ligase ligand.
- E3 Ligase Ligand: A segment that calls up for the presence of E3 ligase to degrade the target protein.
The problem lies in making a PROTAC with the required characteristics. For instance, a linker must allow for proper interaction between the target protein and E3 ligase, but it should not lose its activity through excessive flexibility.
Machine Learning: Changing the Game With PROTAC Design
Enormous datasets can be analyzed by ML algorithms for pattern detection and relationship establishment. Meanwhile, in terms of PROTAC design, this power is exploited by scientists to:
Optimize Linker Design: ML models that study already available data on PROTAC can also analyze how linker structure influences its effectiveness. This provides an opportunity to make linkers more likely to facilitate successful target protein degradation.
Identify Promising Warheads: Through analysis of protein-protein interactions, ML has the potential to identify warheads that could theoretically bind with specific targets.
Predict PROTAC Activity: Researchers can now concentrate on developing a model for predicting the potency and degradation efficiency of a possible PROTAC design using ML, hence aiding in selecting the best candidates for further optimization.
There are two main approaches used in applying ML for PROTAC design:
- Modular Approaches: These models focus on optimizing individual components, particularly the linker. Training data is usually derived from small molecules whose complexity may not fully represent those of PROTACs.
- Comprehensive Approaches: These more advanced models aim at creating the entire molecule, including the E3 ligase ligand and warhead. Although still in their infancy stage, they hold great promise for the future development of PROTACS.
Diving Deeper: The Toolbox of ML-Driven PROTAC Design
What tools are scientists currently using to realize this ‘potential? Let’s take a closer look at some of the main actors in this revolution:
Generative Models:
Imagine that you had a tool that could come up with wholly new molecules based on existing data. This is precisely how generative models work! But what are they good at?
Linker Optimization: Generative models identify patterns in linker design by analyzing successful PROTAC structures. As such, these algorithms generate novel linker structures with desirable characteristics like optimum length and flexibility. Therefore, researchers can explore beyond the readily available space for chemical designs and possibly find more effective linkers.
De Novo PROTAC Design: Some advanced generative models are still in their infancy but have been explored for designing entire PROTAC molecules, including the warhead and E3 ligase ligand. This could be a convenient method for developing PROTACs on a large scale.
PROTAC design has examples of generative models such as:
- Variational Autoencoders (VAEs): These models can learn compressed versions of existing PROTAC structures and initiate new variations based on acquired knowledge.
- Junction Tree Variational Autoencoders (JT-VAEs): This kind of VAE is built especially for molecules, which are represented as graphs where nodes are atoms and edges represent bonds. This makes it possible to capture intricate relationships among different parts of a PROTAC molecule.
Graph Neural Networks (GNNs):
Think about this in terms of a network that not only analyzes individual parts but also understands how they relate to each other. That is what GNNs do! The models perform exceptionally well in:
Understanding PROTAC Interactions: The linker, warhead, E3 ligase ligand, and target protein all interact extensively to enable the proper functioning of a PROTAC. GNNs can analyze these interactions by representing the PROTAC as a graph with nodes that signify components and edges representing connections. Changes made in one part may affect the whole structure of the molecule favorably or adversely, according to analysis conducted by GNNs.
Predicting PROTAC Activity: GNNs can be trained by identifying the intricate interaction within a PROTAC that can then predict how well-given designs would work about degrading the targeted protein. The technique helps in sorting out the most potential candidates for future appraisals.
Some of the specific cases of using GNNs in PROTAC design are:
- Gated Graph Neural Networks (GGNNs): These GNNs contain a gating mechanism that enables them to concentrate on only relevant information within a PROTAC graph, leading to accurate predictions.
- Convolutional GNNs (ConvGNNs): These types of GNN utilize convolutional layers just like those used in image recognition, which allow them to extract local patterns from a PROTAC graph; it is particularly important when explaining how alterations in linker structure could influence its bonding with the target protein.
Reinforcement Learning (RL):
Think about programming your computer to learn through experience, just as you do when training a dog! This is what RL is all about. Using this method for PROTAC design involves:
Linker Optimization: Using RL, a PROTAC and its target protein can be simulated to generate algorithms. Successful target degradation results in rewards, whereas failures lead to penalties. With time, the model will thus learn how to alter the linker structure for effectiveness optimization.
PROTAC Design Exploration: Through RL, the pool of possible PROTAC designs within a chemical space is large. This means that the model can generate a design featuring high target degradation efficiency and low toxicity.
However, even though it is still in its early stages, RL has shown promise in shaping up and speeding up the discovery of optimal candidates for PROTAC. These are only examples among many tools used by researchers to harness ML for designing PROTACs. With these methods and surpassing current data limitations, we are paving the way toward future therapeutic strategies that would make targeted protein degradation the cornerstone of treatment options for a wide range of diseases.
Future is Bright: Join the Talk!
The union of ML and PROTAC design offers an enormous opportunity for revolutionizing drug discovery. As we address the data limitations and incorporate 3D structural information, even more sophisticated ML models can design effective and specific PROTACs. This could result in the birth of completely novel therapeutic strategies against a whole range of diseases.
ML-driven PROTAC design field is moving at a high speed and is very captivating. By collaborating, researchers, clinicians, and data scientists can fully unlock the potential behind ML-driven PROTAC design to achieve a future where targeted protein degradation is a powerful tool in our struggle against diseases.
What are your biggest hopes for ML-driven PROTAC design? Are there specific diseases you’d like to see tackled with this approach? Leave a comment below and share your thoughts!
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.