Scientists from McGill University, Genentech, and Université de Montréal have developed GFlowNets, a new way to speed up the process of discovering drugs. Their findings show how one can now generate new drug molecules by considering their impact on cell morphology.
The Challenge of Drug Discovery
The traditional approach to drug discovery is often a long and costly one that starts with the identification of a protein target, followed by the development of a molecule that will bind to it. This can be time-consuming and, in most cases, achieves nothing. In response, scientists have started using phenotypic screening instead, which looks at compounds affecting an entire biological system rather than targeting individual molecules.
One very important tool used in phenotypic screening is high-content imaging (HCI), which gives comprehensive information on cellular structure and function. By looking at these images, scientists can identify chemical substances that elicit specific changes in cell shape, thereby having potential as therapeutic targets for novel medicines. Nevertheless, designing new drugs based on these morphological changes has been a major problem.
A Fresh Approach: GFlowNets and Multimodal Learning
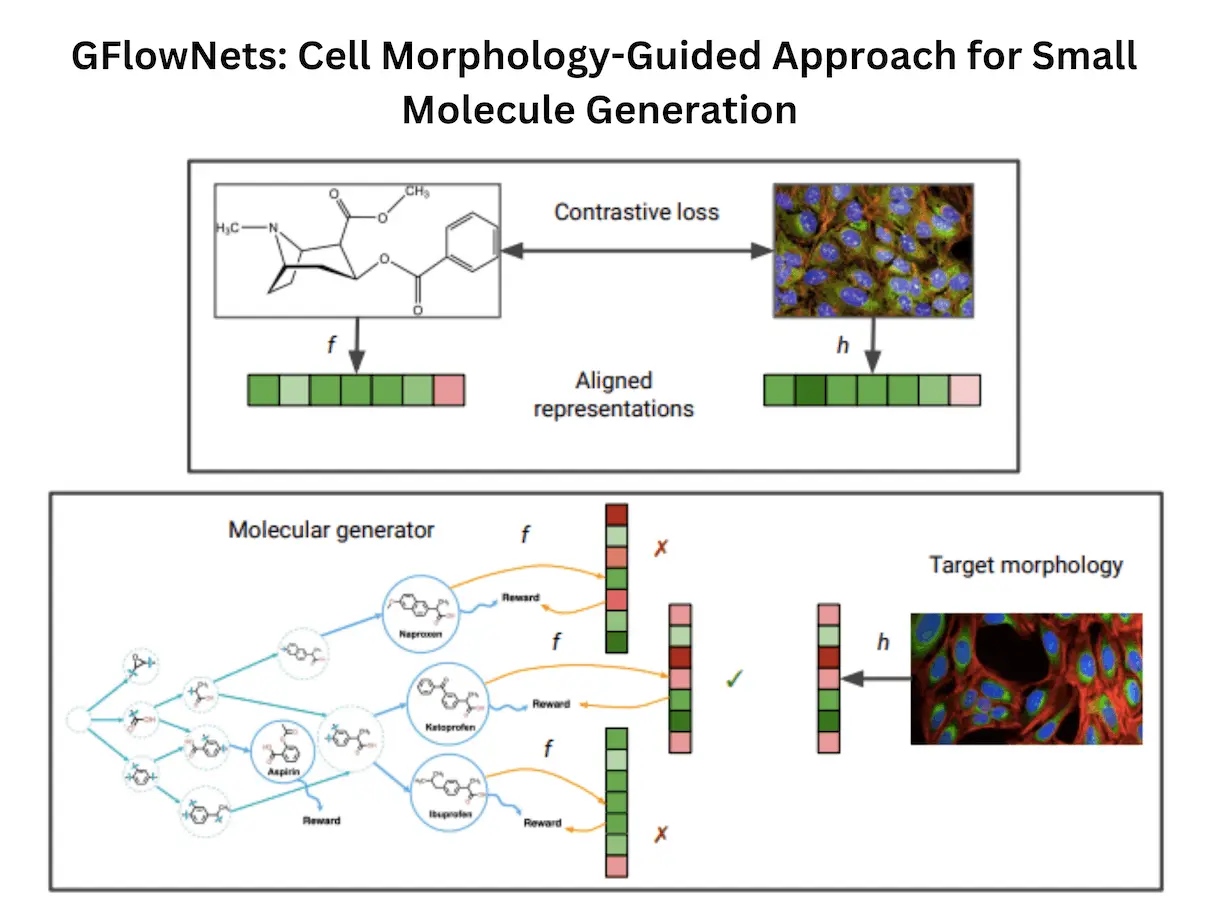
The researchers addressed this problem through the use of two potent methods, namely Generative Flow Networks (GFlowNets) and multimodal contrastive learning. GFlowNets are machine learning models that generate new data samples from a given set of examples. In this case, scientists were generating new molecules.
Multimodal contrastive learning is another machine learning technique in which computers can learn connections between different types of data, like molecular structures or images. Therefore, the authors applied it to cell images together with molecular data to come up with a model that connects specific cell morphologies with the compounds that brought about those morphologies.
How It Works
The multimodal contrastive learning model is first trained on a large dataset of molecules and corresponding cell images. This gives the model an understanding of particular cell morphological patterns associated with certain molecular structures. Then, one could input any image into this trained model to get its “latent representation.” This representation simply puts down some numbers determined by some features of the picture being used.
Furthermore, to achieve this goal, GFlowNet is trained to generate molecules that have latent representations that are similar to the target cell image. Essentially, the model learns how to create molecules that would most likely produce desired morphological changes. Thus, scientists can identify good candidates by continuously generating and refining molecules.
Step 1: Making Connection
- Multimodal Contrastive Learning: To envision how to teach a computer the connection between apples and reds is basically what the multimodal contrastive learning model does. It gets adept at relating definite molecular structures (apples) to changes in cell shape (red).
- Generation of Code: When trained, the model assigns a unique code or ‘latent representation’ to each cell image. This code serves as an identity card that stores the salient characteristics of the appearance of the cell.
Step 2: Designing New Molecules
- GFlowNets: A creative machine is what GFlowNets is. In this case, it’s used to create new molecule structures.
- Matching the Codes: The digital fingerprint of molecules should be closely related to their “latent representation,” which belongs to any desired form from cells among molecules. For example, finding a molecule that ‘looked’ similar in computer vision terms to the target cell image.
- Iterative Improvement: This step continues refining the generated molecules until they are very close approximations of the required cells’ morphology.
This new approach to drug discovery could change everything by providing a faster and more targeted way of discovering new drugs. Instead of focusing on the molecular target but rather on what they want (cell morphology), researchers can investigate a wide range of possibilities and increase their chances of finding potential therapeutic agents.
Potential Impact
While still early in its stages, this research has shown some promise. They demonstrated the ability of their method to produce molecules with high similarity to known drug-like compounds, indicating that such generated molecules might be viable drug candidates. In future years, we will witness the emergence of even more sophisticated techniques for drug discovery as artificial intelligence technology continues to evolve. Scientists are thus converging computational prowess with biological insights, moving us towards a world where better therapies can be developed faster than ever.
Conclusion
The union between artificial intelligence and biology and exciting advancements in drug discovery are happening. By utilizing such advanced techniques as GFlowNets and multimodal contrastive learning, researchers are making significant leaps in their attempts to design molecules with desired properties. This research represents a positive step towards hastening the process of developing effective treatments for various types of diseases despite still facing challenges. Even more groundbreaking advances will be experienced within this field as technology continues to evolve.
This is an exciting breakthrough in drug discovery! What are your thoughts on this new approach? Do you think it will revolutionize the pharmaceutical industry?
Share your insights and questions in the comments below!
Let’s start a discussion!
Article Source: Reference Paper | Code available on GitHub
FAQs
A: GFlowNets refers to a machine-learning model that can create new data samples from given sets of examples. In this case, it was used to generate new drug molecules.
A: Multimodal contrastive learning is a technique in machine learning that causes computers to learn relationships between differing data types, such as images and molecular structures.
A: This research is still at its nascent stage; hence, while findings look promising, they will require time before they translate into real-world drug development.
A: No, the approach caters to human scientist assistants rather than replacing them.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.