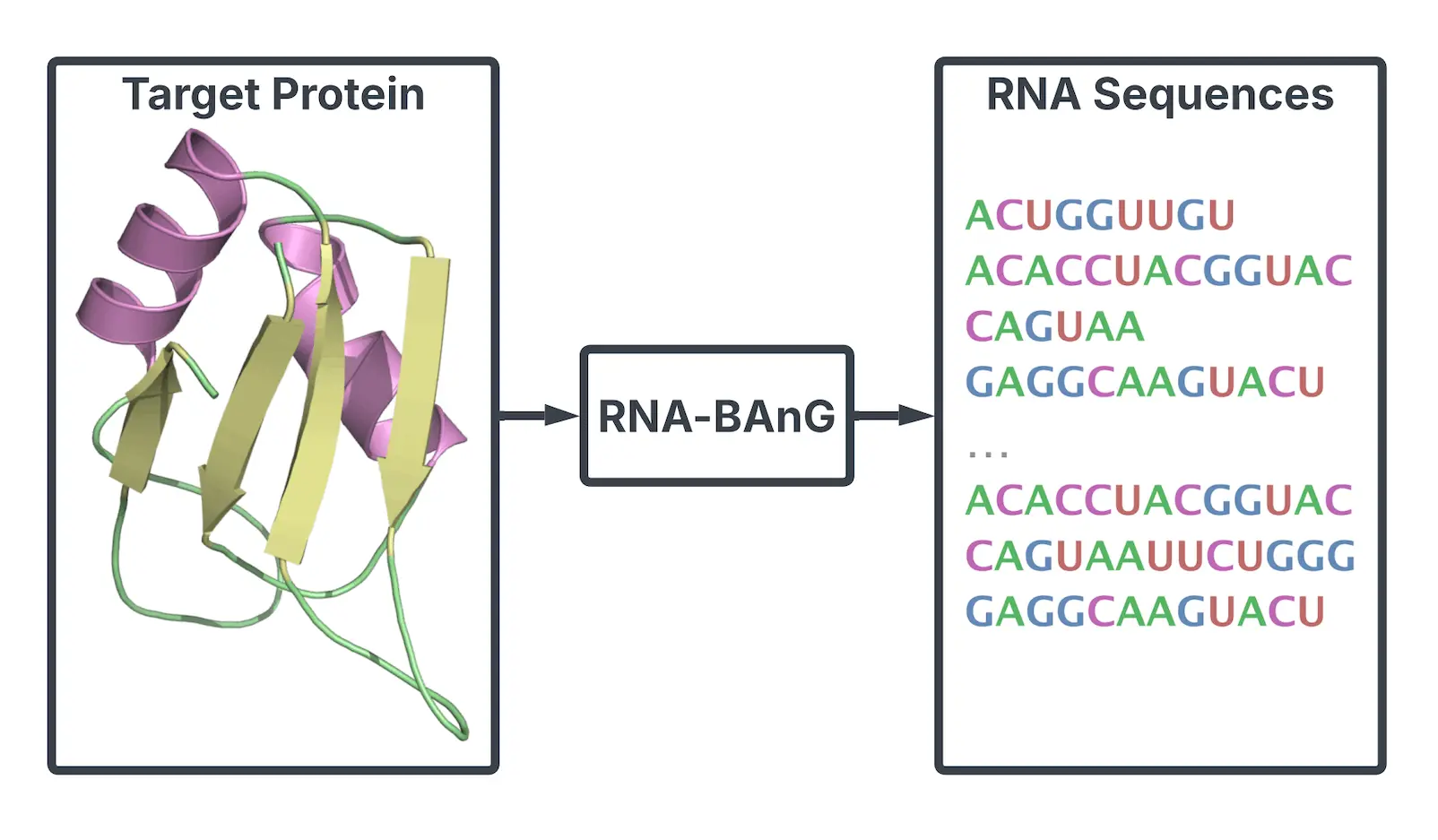
One of the most significant challenges in computational and experimental biology is creating RNA molecules that interact with particular proteins. The practical use of current computational methods is limited by their requirement for a significant number of empirically confirmed RNA sequences for every individual protein or a thorough understanding of RNA structure. Researchers have developed RNABAnG, a deep learning-based model that can produce RNA sequences for protein interactions without these prerequisites, to overcome this constraint. The strategy is based on a novel generative technique called Bidirectional Anchored Generation (BAnG), which takes advantage of the finding that functional binding motifs are frequently contained within larger sequence contexts in protein-binding RNA sequences.
Introduction
Deep learning has greatly benefitted structural and synthetic biology by improving protein design. Applications in synthetic biology and drug discovery have been made possible by this technology’s increased efficiency in macromolecular design. Significant advancements have been made in protein sequence design, including ESM3 and Chroma. This has created new avenues for tackling intricate biomolecular problems, such as RNA synthesis, where functional RNA sequences can be designed using deep learning techniques.
Creating RNA sequences that can attach to particular proteins is a crucial endeavor known as “RNA design.” A prominent example of such molecules is aptamers, which are short sequences of single-stranded RNA. They have a wide range of uses in diagnostics and therapy because of their high affinity and specificity for particular proteins. Aptamers are traditionally found by systematic evolution, which is a time-consuming experimental procedure. The discovery of aptamers might be sped up and made simpler by developing computational techniques for their design, which would increase their potential for use in biomedical applications.
Numerous studies on RNA generation have focused on proteins using various models and tools. Diffusion processes and flow matching have been explored in recent studies such as AptaDiff and RNAFLOW, but these approaches rely on a large collection of nucleotide sequences, which limits their applicability to proteins with extensive experimental data. RNAFLOW, the only method not trained on RNA affinity experimental data, relies on RNA structure prediction tools to guide RNA design, which frequently lacks the accuracy required for precise RNA structures.
RNA-BAnG
In this study, scientists provide a deep learning model and a novel generative approach that doesn’t require RNA structural knowledge or particular experimental data for the target protein to function. Compared to the previously listed methods, this strategy allows for improved efficiency and a wider range of applications in the synthesis of RNA sequences. The transformer construction with geometric attention forms the basis of the model, which is called RNA-BAnG. The latter makes it possible to incorporate knowledge about protein structures, which is essential for forecasting interactions between RNA and proteins. Even in cases where the target protein cannot be solved experimentally, researchers can still collect extremely precise structural data by using AlphaFold2, a state-of-the-art protein structure prediction method. Utilizing the sequence and structural characteristics of a particular protein, the RNA-BAnG model and the generative approach generate RNA sequences that interact with it.
Motivation for the Study
The inspiration for the design of the proposed generative method arises from two key observations. First, it is frequently unclear how long the total RNA sequence produced will be. Consequently, an autoregressive approach is used in the procedure. Second, functional binding motifs, or particular areas that mediate interaction by establishing molecular interactions with the protein are present in most RNA sequences that interact with proteins. Because these binding motifs are integrated in broader sequence contexts, the binding specificity is less affected by the nearby non-binding sequences. This means that, as is frequently done in the most advanced NLP autoregressive models available today, it is more efficient to start sequence production from the binding motif rather than the ends of the sequence. These are the basis of the Bidirectional ANchored Generation (BAnG) technique.
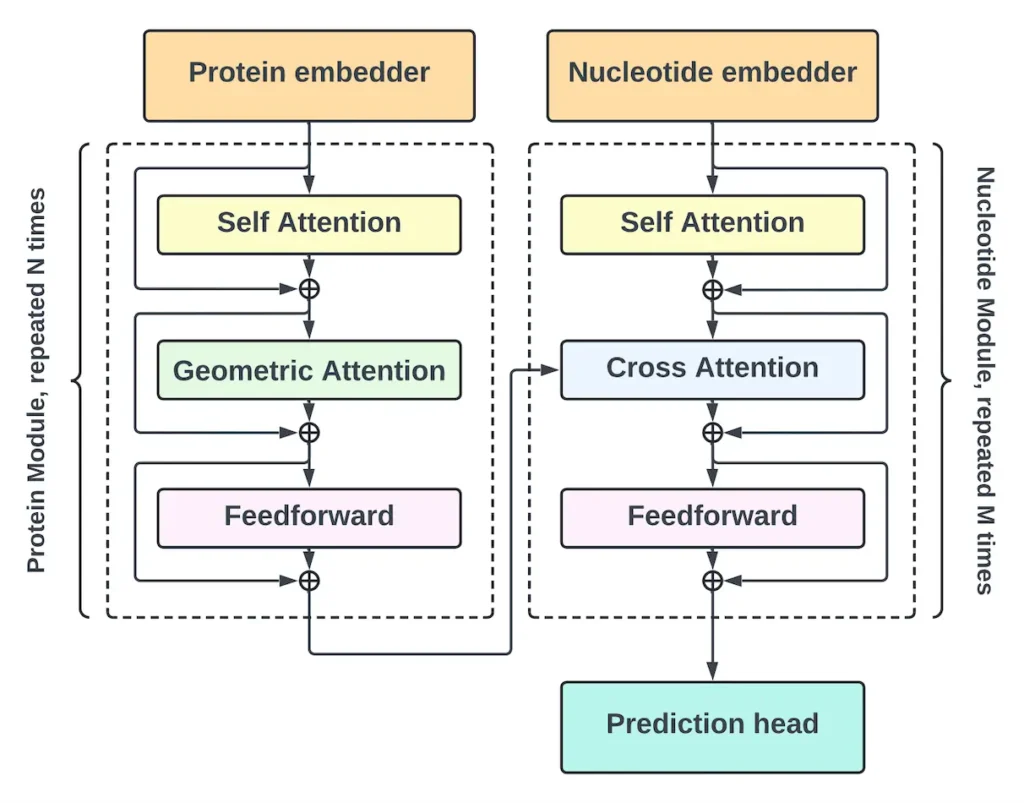
Architecture of RNA-BAnG
The protein module and the nucleotide module make up the two primary parts of the RNA-BAnG architecture. While the nucleotide module creates a nucleotide sequence conditioned on this representation, the protein module uses the structure and sequence of the protein to create a representation. The modules of the model are composed of the following primary building blocks: Cross Attention, Embedder, Self Attention, and Geometric Attention. Token embeddings are created from DNA, RNA, and protein sequences by the Embedder block. Because it can assist the model in learning common patterns, the addition of RNA and DNA sequences to the training data enhances the dataset.
Future Work
The model’s design could be optimized for better efficiency, its usability for wider applications could be improved, and experimental feedback might be incorporated to refine the model in future work. Experimental confirmation of the produced sequences would also shed more light on the usefulness of the approach in real-world scenarios. All things considered, the work is a major advancement in the creation of RNA sequences and provides researchers with a strong and versatile tool for studying RNA-protein interactions.
Conclusion
A new deep-learning model called RNABAnG and a sequence generation technique called BAnG are presented in the paper for creating RNA sequences that bind to proteins. There is no need for a lot of structural or interaction data with this flexible approach. Protein structure is necessary, yet sequence information is adequate according to AlphaFold predictions. Because RNA sequences have functional binding motifs, the surrounding sequence context is less important for interaction. This discovery forms the basis of the technique. Beyond RNA-protein interactions, BAnG is useful for optimizing functional subsequences inside longer sequences and showed exceptionally well in synthetic tasks. By producing a greater percentage of sequences with significant projected binding affinity, RNA-BAnG performs better than current techniques, according to DeepCLIP scoring.
Article Source: Reference Paper | The code and the model are not yet released.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.