In the past few years, there has been a significant increase in multi-omics data generation for biomedical research. However, the analysis of these large-scale, complex, and heterogeneous datasets represents an enormous challenge for researchers. Recently, researchers from the Allen Institute of Immunology have developed a ground-breaking PALMO platform, which contains five analytical modules allowing the examination of longitudinal multi-omics data – from bulk or single-cell samples – from multiple perspectives. The modules are, in turn, responsible for the breakdown of sources of variability within the data, the collection of stable or variable features across time points and for participants, the identification of biomarkers across time points of each participant, and the investigation of outliers. Together, these investigated parameters are indicative of disease mechanisms and treatment responses, allowing for improved personalized medicine and treatments for patients.
Longitudinal Multi-Omics Data, its Significance, and the Need for Improved Methods to Analyze It
Longitudinal multi-omics refers to information about different types of biomolecular data, like nucleic acids (DNA and RNA), proteins, and metabolites, as a function of time for specific samples. This means that a single study could include data about the transcriptome, protein levels, epigenetic alterations, genomic mutations, and glycomics. By combining all these individual molecular features and data, scientists could obtain a complete picture of the biological system in bulk tissue or single cells, which can help them understand disease progression over time. However, analyzing these datasets is not a straightforward task. This is because the data from individual omics platforms must all be processed, integrated, and analyzed. In addition, each platform would have its own limitations and biases; therefore, it is not simple to address the limitations and biases of each omics platform altogether. Lastly, the need to develop normalized and standardized methods for analyzing longitudinal multi-omics data (regardless of human heterogeneity) is yet to be fulfilled. Collectively, these challenges illustrate the need for robust and suitable analytical tools to process and analyze complex longitudinal multi-omics data.
The PALMO Platform and its Uses
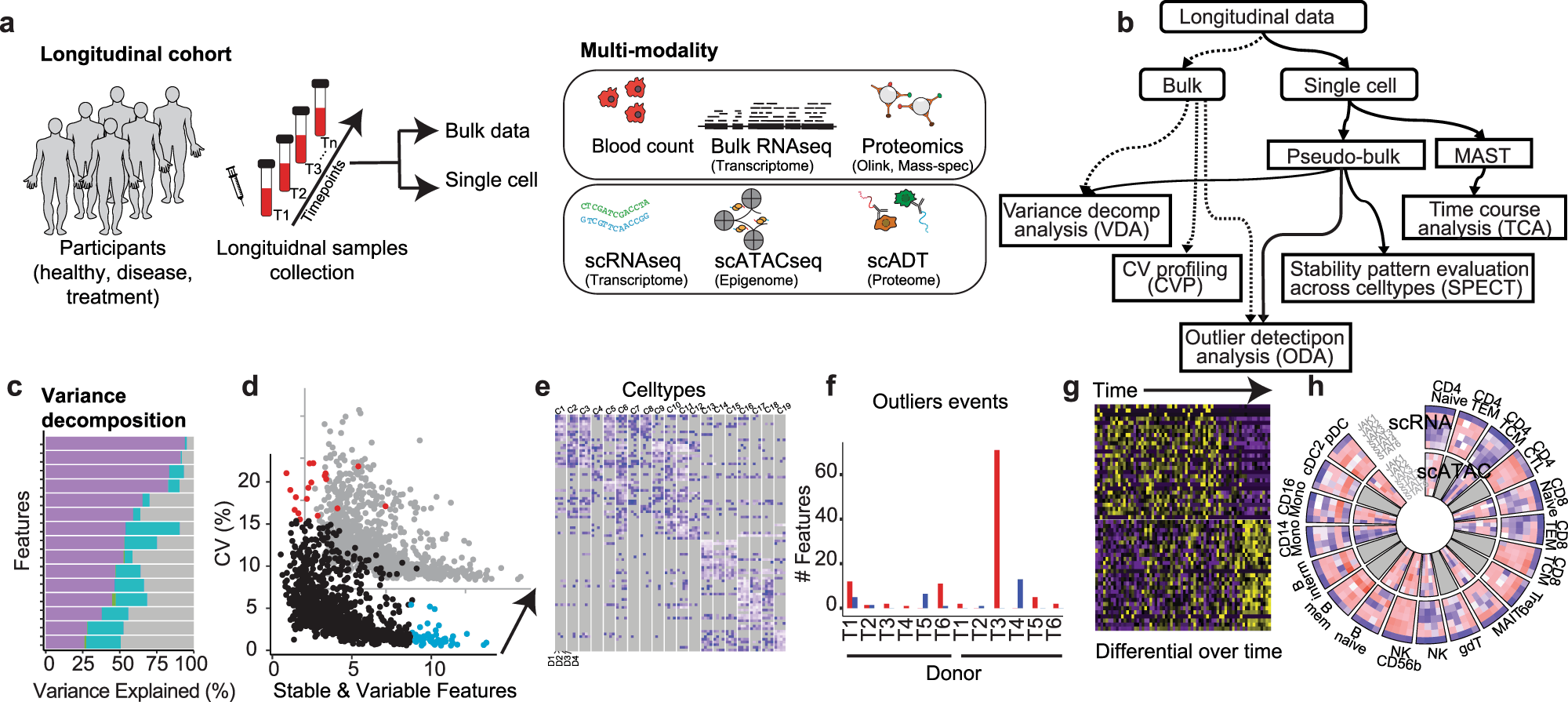
Researchers’ recent development of a novel platform – PALMO – appears to be an appropriate and accurate solution for analyzing longitudinal multi-omics data by addressing the mentioned challenges. PALMO is a free and easy-to-use software package that thoroughly simplifies the processing and analysis of longitudinal bulk and single-cell omics data. It provides five analytical modules – variance decomposition analysis (VDA), coefficient of variation profiling (CVP), stability pattern evaluation across cell types (SPECT), outlier detection analysis (ODA), and time course analysis (TCA), offering crucial information from various perspectives. Several aspects of PALMO allow it to address the challenges of analyzing longitudinal omics data. Firstly, it is easy to use because it offers a straightforward and guided user interface (UI) that does not require any advanced skills to use. Moreover, PALMO also produces visual data, like circus plots, which can augment data processing.
Lastly, because it provides a range of analytical modules, researchers can evaluate different features of the longitudinal omics data. The five modules allow researchers to gain insights from longitudinal multi-omics data – which includes transcriptomics, genomics, epigenomics, glycomics, proteomics, and metabolomics – in a more direct and simple manner.
The five modules and their functions;
1. VDA: This module evaluates the contributions of biomolecular factors of interest to the total variance of individual features.
2. CVP: This module assesses the intra-participant variation over time in bulk data and identifies consistently stable or variable features among participants.
3. SPECT: This module assesses longitudinal stability patterns of features in single-cell omics data and identifies stable or variable features unique to individual cell types but consistent among participants.
4. ODA: This module examines the possibility of abnormal events occurring during a longitudinal study.
5. TCA: This module evaluates the transcriptomic changes over time based on longitudinal scRNA-seq data of the same participant and identifies genes exhibiting significant temporal changes.
Image source: https://doi.org/10.1038/s41467-023-37432-w
The Effectiveness of PALMO
The performance and effectiveness of PALMO were tested on longitudinal omics data from various sources and using various technical methods, including clinical lab test results, bulk or single-cell omics, various TCR sequencing data, and information on gene expression levels, the abundance of proteins, and cell type composition. Basically, PALMO was assessed using different datasets and compared with other methods/tools. The results in the paper clearly show that PALMO outclassed other platforms in analyzing longitudinal multi-omics data.
The paper describes a complex longitudinal multi-omics dataset that was used to illustrate the effectiveness of PALMO. The dataset included six healthy, non-smoking Caucasian donor blood samples over a 10-week period. Five data modalities were included – complete blood count, high-dimensional flow cytometry, plasma proteomics, droplet-based-scRNA-seq, and droplet-based scATAC-seq. Then, the performance of each of the five modules was assessed and compared to other tools, and the results were quite remarkable. For instance, the VDA module, which is used to evaluate the contributions of factors of interest to the total variance of individual features, was assessed based on how it evaluated the inter-donor and intra-donor variations in bulk data (e.g., the plasma proteomics data) based on the specific donor and timepoint as factors of interest. The results demonstrate that inter-donor variations accounted for more than half of the total variance.
Moreover, VDA was also used for scRNA-seq and scATAC-seq data, where the cell type was the factor of interest. The results for this demonstrated that inter-cell-type variations were far more prominent than inter-donor and intra-donor variations in the two single-cell data modalities. In fact, some of the genes had more than 50% of their total variance from inter-cell-type variations. These findings highlight the prowess of VDA and how it can be used as an effective tool for analyzing multi-omics data and, more specifically, identifying sources of variations in the data.
Similarly, all other individual modules of PALMO were assessed based on their individual functions, and the results were exemplary. Overall, the results indicate that PALMO is robust and accurate and can be used for longitudinal multi-omics analysis in order to provide critical insights into the underlying biomolecular mechanisms of significant processes, including diseases. It is also worth noting that the researchers observed that PALMO could detect ‘STATIC’ genes (genes that do not change over time), which could act as markers to differentiate between cell types and provide information on biological differences.
Lastly, when PALMO was compared with other tools like TCR, variancePartition, and Seurat, PALMO could work better in combination with TCR. Nevertheless, PALMO showed better results (dynamic changes) than Seurat.
Conclusion
To conclude, PALMO can be a revolutionary tool for researchers working on longitudinal studies who need to analyze large and complex multi-omics datasets. Its flexibility based on the five modules, ability to handle missing data, and user-friendliness make it stand out from other tools.
Article Source: Reference Paper
Learn More:
Diyan Jain is a second-year undergraduate majoring in Biotechnology at Imperial College, London, and currently interning as a scientific content writer at CBIRT. His passion for writing and science has led him to pursue this opportunity to communicate cutting-edge research and discoveries engagingly to a broader public. Diyan is also working on a personal research project to evaluate the potential for genome sequencing studies and GWAS to identify disease likelihood and determine personalized treatments. With his fascination for bioinformatics and science communication, he is committed to delivering high-quality content a CBIRT.