Researchers from the University of Shanghai, China, created PackDock, a diffusion-based side-chain packing model that predicts diverse side-chain conformations in protein binding pockets, both with and without ligands. PackDock is compatible with other docking algorithms and achieves near-rigid docking accuracy while being significantly faster than traditional docking methods.
The Promise and Limits of Drug Design Using Protein Structure
Structure-based drug design (SBDD) has been a central strategy for drug discovery for decades, relying on accurate protein structures to design ligands that bind effectively. In simplest terms, SBDD depends on knowing how proteins and ligands fit together.
Techniques like X-ray crystallography, cryo-EM, and computational tools, such as AlphaFold, have made 3D protein structures more accessible than ever. But the protein structures we get from these methods are usually static snapshots. They don’t capture the dynamic flexibility of proteins, especially failing to reflect how they internally shift to accommodate ligands via conformational selection (the ligand chooses from pre-existing protein conformations) or induced fit (the protein changes shape after the ligand binds) or both.
A traditional solution to this problem is Molecular Dynamics simulations, but they are computationally expensive and too slow for large-scale virtual screening.
Beyond Lock and Key for SBDD: Strategy for Flexible Docking Instead of Rigid Docking
Instead of modelling full protein flexibility, a practical strategy is to allow local flexibility in the binding pocket. This balances accuracy with complexity. Existing approaches like flexible docking using Vinaflex, IFD-MD, or deep-learning-based methods are either not scalable, computationally expensive, or fail to provide atomic-level details needed for SBDD.
We require a flexible docking method that:
- Efficiently models protein side chain conformational changes
- Avoids pitfalls of current deep learning approaches
- Produces atomic-level, physically realistic complexes
Introduction to PackDock: A Practical Approach for Flexible Docking
PackDock is introduced as a flexible docking pipeline, which is a hybrid of both conventional docking methods (rigid/flexible) and deep learning based methods (generative models). It also integrates the two classical theories of binding discussed above.
It can also capture critical amino acid conformational changes that are important for ligand binding. This ability provides guidance for lead optimization, since drug designers can see which residues shift and adapt during binding.
Understanding PackPocket: A Two-Stage Pipeline
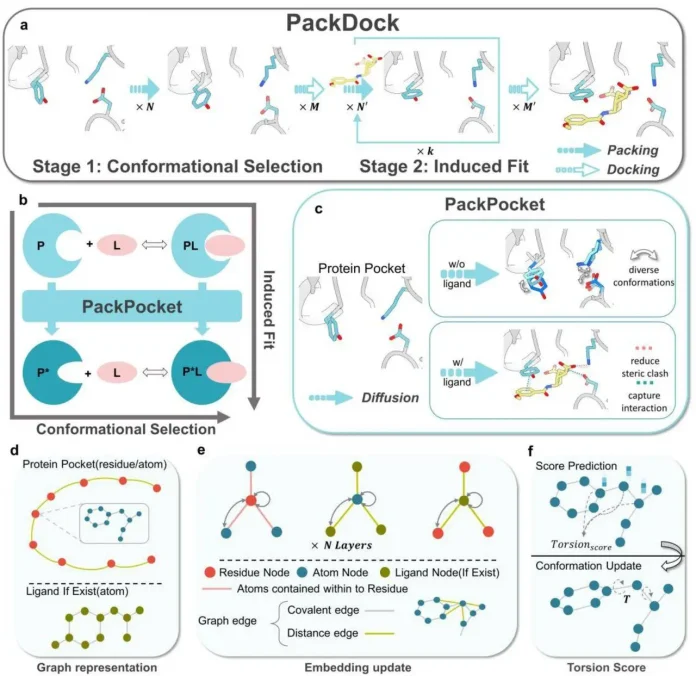
The core innovation behind PackDock is PackPocket, a diffusion-based model that predicts torsional angles of side chains in binding pockets. This allows it to model conformational changes in Free (apo) states and Ligand-bound (holo) states. By focusing on torsional angles, PackPocket simplifies the complex problems of side chain flexibility modeling. The pipeline operates in two stages:
Stage 1: Conformational Selection
During stage 1, PackPocket explores side chain orientations in the empty binding pocket. This simulates the conformational selection hypothesis, where ligands bind to pre-existing receptor conformations. This gives a wide range of receptor conformations ready for docking as an output.
Stage 2: Induced Fit
At this stage, the conformations from stage 1 are used as receptors for docking. The ligand conformation obtained during the docking process is then used to model how the ligand influences pocket side chains. This simulates the induced fit hypothesis, where the receptor adapts after ligand binding, resulting in dynamic receptor-ligand complex conformations as outputs.
Furthermore, PackPocket’s module relies upon graph representations, which is one of its most important innovations, as it allows the model to capture protein ligand interactions at atomic levels while still being computationally efficient.
It uses Atom Graph (Protein Pocket Region) and Ligand Graph (if present). In an atom graph, each residue in the binding pocket is represented as a residue node, and within each node are atom nodes for every atom, while in a ligand graph, each ligand atom itself is treated as a node.
How PackPocket is Validated Across SBDD Tasks
When tested on real-world drug discovery scenarios, side-chain packing and re-docking validate that PackPocket shows superior accuracy in predicting side-chain orientations and ligand positions when docking back into known structures, outperforming existing methods (SCWRL, FASPR, AttnPacker, DiffPack).
Other than this, PackPocket showed strong performance in cross-docking (apo to holo) tests, docking into apo (unbound) or non-homologous holo structures, which reflects realistic drug design challenges.
Moreover, Hypothetical models demonstrated that PackPocket could even start from predicted protein structures like those from AlphaFold, enabling SBDD without experimentally determined structures.
Key Takeaways
PackDock is a hybrid diffusion-based flexible docking pipeline that efficiently models side-chain flexibility, blends classical binding theories, and produces atomic-level accuracy across real-world drug design scenarios, eliminating the gap between rigid docking and MD simulations.
Article Source: Reference Paper | Published Abstract in PNAS | Code Availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.















