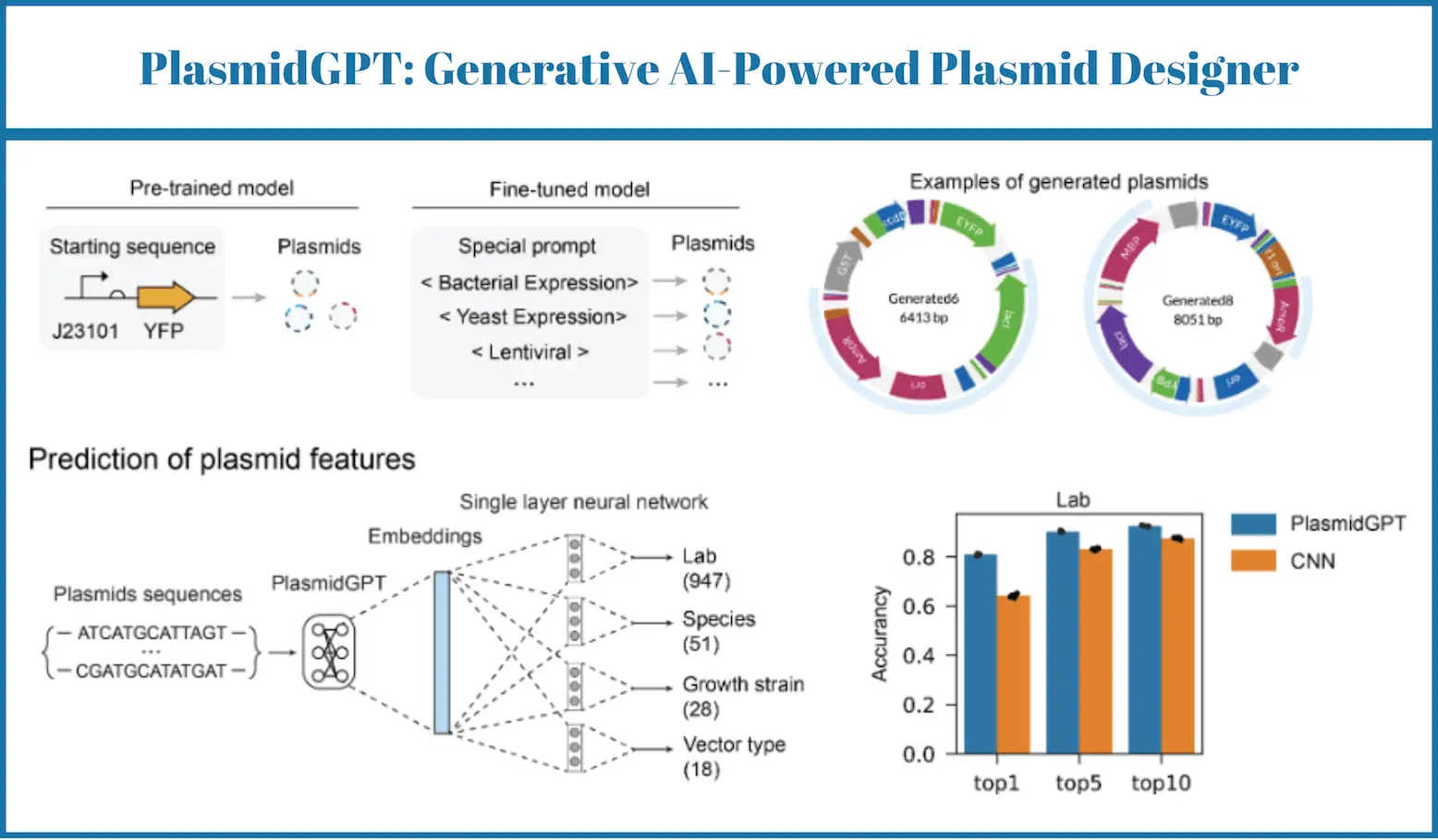
Recent research shows that within the domain of synthetic biology, there have been tremendous improvements, especially in designing and engineering biological systems. Plasmids, crucial tools in genetic manipulation, play a pivotal role in this revolution. However, the traditional way of designing and adding notes on the plasmids is tedious and time-consuming. To solve this problem, a team of researchers developed PlasmidGPT, which helps in the design and analysis of plasmids by automating processes.
Understanding Plasmids
Plasmids are relatively small, generally circular structures of DNA that are present in bacterial cells and a few other microorganisms, too. These are dissimilar from the main chromosomal DNA and can be self-replicated. Plasmids are influential in the field of genetic engineering for their complex vectors that can retain and replicate extra-chromosomal elements. They are often employed as a means of DNA delivery, synthesis of requisite proteins in a subject organism, alien DNA fragment replication and expression, and even genome editing.
The Limitations of Traditional Plasmid Design
The traditional approach to plasmid design involves several steps, including selecting appropriate genetic elements, assembling them into a desired construct, and verifying the accuracy of the sequence. This procedure, however, has limited applicability because it is very long, requires much effort, and may not always be accurate. Traditional plasmid design suffers some of the most obvious problems, such as small design space, long turnaround time, human error, and non-standard processes. These limitations have prompted researchers to seek better plasmid design methods, hence brand new technology like PlasmidGPT.
The Power of PlasmidGPT
Researchers who developed PlasmidGPT offered us a breakthrough. This generative language modeling approach is more useful as it is incorporated with a dataset of various plasmid sequences.
Key Features of PlasmidGPT
De Novo Plasmid Generation: PlasmidGPT is capable of inducing novel plasmid sequences that are similar to known ones but exhibit low sequence identity to the training data. This allows functional plasmids with new designs to be obtained within a very wide design space.
Conditional Plasmid Generation: The model is capable of plasmid generation based on specific input sequences or design constraints, providing greater control over the final product. For example, researchers can specify the desired genetic elements or target organisms to obtain customized plasmids.
Plasmid Annotation: PlasmidGPT can annotate the genetic elements into the plasmid sequence, such as the promoters, ribosome binding sites, coding sequence, and replication origin. This particular type of information is critical in the understanding of the function of plasmids and their improvement for other uses.
Prediction of Plasmid Attributes: The model can predict some of the attributes of genetic parts within plasmid sequences, identifying elements such as promoters, ribosome binding sites, coding regions, and replication origins. This can help researchers avoid potential compatibility problems and design an appropriate plasmid for a desired end application.
Uses of PlasmidGPT
There are a plethora of possibilities for the PlasmidGPT applications in:
- Synthetic Biology: Researchers can use PlasmidGPT to rapidly design and construct novel genetic circuits and biosensors.
- Biotechnology: This model can help speed up therapeutic protein development, vaccines, and biomaterials.
- Bioinformatics: PlasmidGPT can be used to analyze large datasets of plasmid sequences, identify patterns, and gain insights into plasmid biology.
Future Directions and Conclusion
At the same time, although PlasmidGPT provides ample improvements in the area of plasmid design and analysis, it can be further enhanced. Possible further work may be centered on the following:
- Improving model complexity: Developing the ability of the model to construct more intricate and active plasmids.
- Expanding vocabulary: Using more gene elements to extend the range variations of the DNA sequences.
- Integrating experimental data: Combining computational models with experimental data to validate and refine predictions.
- Developing user-friendly interfaces: Making the interface of such tools less complicated for nonspecialists to successfully apply them.
PlasmidGPT is an exceptionally powerful software capable of changing the outlook of the design and analysis of plasmid vectors. PlasmidGTP has the potential to increase the speed and reduce the costs incurred in carrying out research and facilitate the invention of remarkable solutions in synthetic biology and biotechnology. As synthetic biology continues to evolve, PlasmidGPT is poised to play a central role in shaping its future.
FAQ
PlasmidGPT is based on the completely embedded use of transformers to encode/decode vector sequences. It can understand and generalize the principles of designing a plasmid, and as a consequence, it can design patterns that belong to known plasmids as well.
Regarding plasmid synthesis, PlasmidGPT can provide satisfactory constructs for various fields such as research, economic exploitation, biotechnological activity, and synthetic biology. However, it is important to note that the generated plasmids may not be fully functional or optimized for specific applications.
Likewise, the performance of PlasmidGPT is directly linked to the quality of the available data that was employed during training and the task of plasmid design that must be accomplished. While the model has been found effective in designing plasmids that have generated their intended plasmids from the described one, the function or performance may not be consistently predictable in any viral or plasmid that has been proposed.
Yes, PlasmidGPT remains available to the general public for research and development purposes. Access to the model and its code was made available on GitHub.
Article Source: Reference Paper | Trained model and inference codes are available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.