RNA, often overshadowed by its protein counterpart, plays an essential role in the molecular machinery of life. Predicting RNA’s three-dimensional (3D) structure is a vital challenge in understanding its function, but it’s a task riddled with complexities. Enter NuFold, an innovative RNA structure prediction method that promises to advance our understanding of RNA folding mechanisms. This groundbreaking research was conducted by a team of researchers from Purdue University, USA, and has opened new possibilities in the realm of RNA bioinformatics.
Why Predict RNA Structures?
The RNA tertiary structure is responsible for carrying out vital functions like gene regulation and synthesizing chemical reactions using ribozymes. Conventional X-ray crystallography or cryo-electron microscopy used for the structural analysis of RNA is expensive and doesn’t completely cover the matter promptly. Although Computational methods are effective and quick, they tend to have low accuracy. Fortunately, NuFold changed the game because it has developed a fast and accurate way to predict the fully atomistic 3D structure of RNA directly from its sequence.
What Makes NuFold Unique?
NuFold has several unique traits, including:
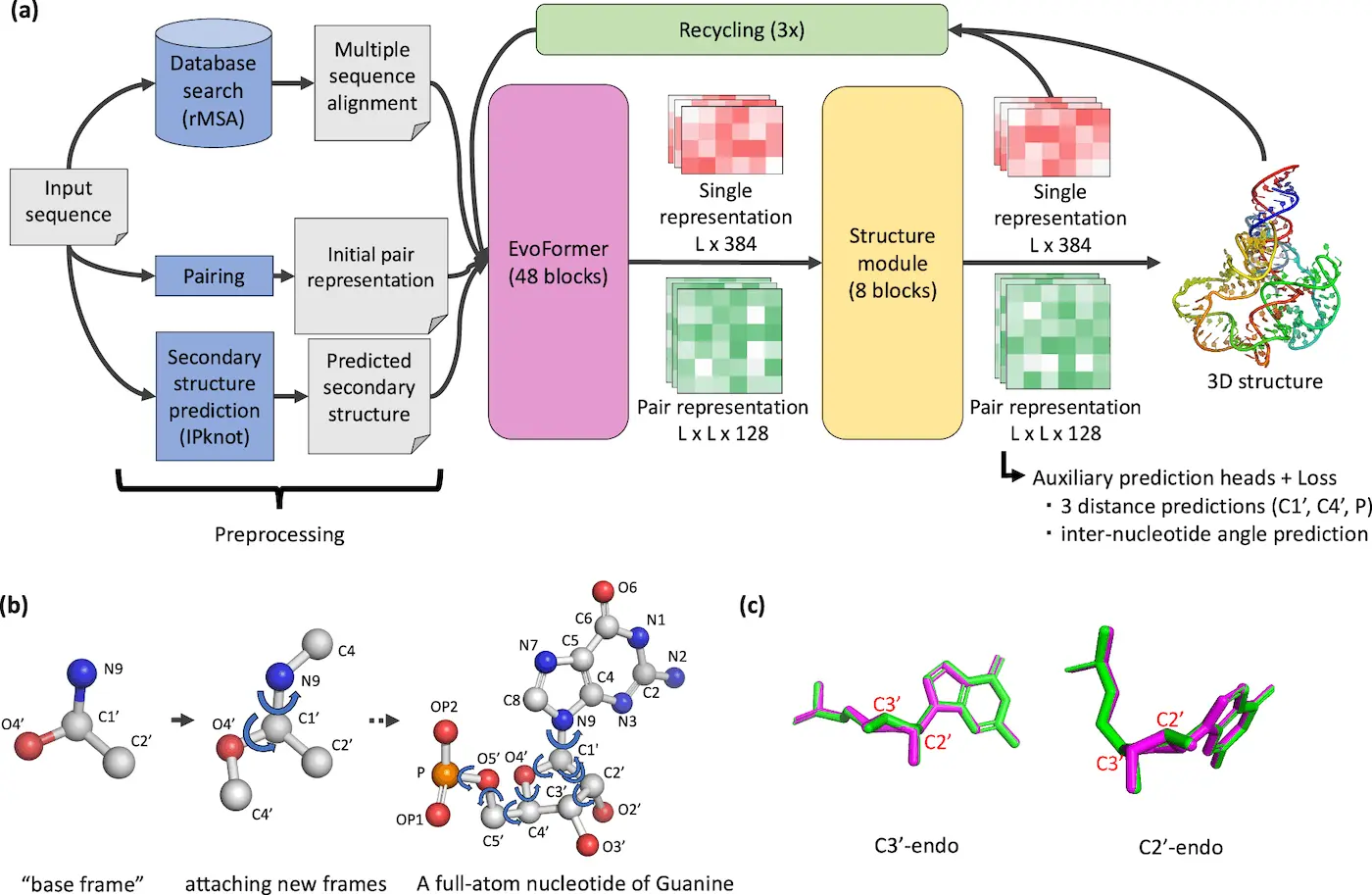
Flexible Nucleobase Center Representation: NuFold’s flexible nucleobase center representation is different from other coarse-grained models. In his approach, NuFold can accurately define the structural motifs of RNA at an atomic level. Such details involve precise control of torsion angle optimization.
Integration of Metagenomic MSAs: The model is provided with MSAs extracted from metagenomic databases to make better predictions. Additional data from RNAcentral, Rfam, and NCBI databases improve the model’s comprehension of RNA’s evolutionary landscape.
Auxiliary Heads for Enhanced Training: The neural network architecture of NuFold also contains extra heads, which enable it to predict other types of structural features. These features include inter-nucleotide torsion angles and inter-nucleotide distances. These enhancements allow the model to more accurately predict the mechanisms of RNA folding.
Breaking Down the Results
The performance of NuFold was tested using tertiary RNA structures from the Protein Data Bank (PDB) and benchmarks from RNA-Puzzles, a community-driven initiative for testing RNA 3D structure prediction models. NuFold was able to reliably reproduce accurately the native RNA structures. For example, the dimerization initiation site of genomic HIV-1 RNA (PDB: 462D) had an RMSD of only 1.71 Å, which shows that it was able to capture RNA’s native fold. In addition, this study also discovered that shorter linker lengths, such as 10 nucleotides, were more accurate. NuFold could pick the best or near best models from the ensemble most of the time about the predicted local Distance Difference Test (pLDDT) scores.
However, there were problems when it came to complex structures. NuFold was able to do well with monomer RNAs but did not do well with some multimeric RNA structures. For example, the hammerhead ribozyme (PDB: rp15) had an RMSD of 14.8 Å because of missed interchain base pairs, which is a place that needs further refinement.
How is NuFold Different?
NuFold had a better range of accuracy than classical energy minimization techniques and fared better than other tools tailored toward RNA prediction with deep learning. The fully atomic model of NuFold was six cases behind its deep learning competition due to insufficient training data. These situations define the need for more extensive databases and better incorporating complementary information such as DMS-MaPseq, SHAPE-MaP, and others to make better predictions.
Conclusion
NuFold holds a monumental significance in RNA tertiary structure prediction. Its unique metagenomic architecture allows NuFold to deeply learn and solve existing problems. Although some work remains, NuFold has made preeminent progress in basic and applied scientific research. The model’s potential to transform the field of RNA biology and therapy keeps coming into clearer focus; therefore, as researchers make additional improvements to the model, its potential only increases.
Ultimately, it appears that RNA is receiving the focus it should have received a long time ago. With NuFolds’s revolutionary architecture at the forefront of the transformation, there is a high chance NuFold could change everything.
Article Source: Reference Paper | The source code of NuFold is available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.