An inter-institutional collaboration across the US, Germany, and the Netherlands introduces MHub.ai, an accessibility and reproducibility-prioritizing, open source platform for standardized access to AI models for automating and accelerating research in medical imaging.
AI as a Revolution in Medical Imaging and Issues with Its Usability
Medical imaging has been a revolution in the medical field for decades, as it allows doctors to detect diseases, plan therapies, and monitor patients without any invasive or painful procedures, as well as before the symptoms worsen, using technologies like X-rays, CT scans, MRI, and ultrasounds to visualize the insides of the body.
However, different radiologists may interpret the same scan differently. This variability can affect diagnosis, treatment, and most importantly, research reproducibility.
AI in medical imaging has already proven it can match, and in some cases surpass, human-level performance. An example can be the unmatched accuracy of Google AI in detecting breast cancer from mammograms, reducing false positives and negatives.
However, AI still hasn’t made it into everyday clinical use as different research groups build models with different architectures, coding styles, and dependencies that are difficult to replicate, making them hard to compare or reuse for non-experts.
Most hospitals and clinics universally use Digital Imaging and Communications in Medicine (DICOM) as the standard format for medical images. That is where the biggest bottleneck is, as most AI models are trained on alternative formats (like JPEG, PNG, NIfTI) and hence, researchers have to manually convert DICOM files into these formats. For large datasets, it’s a time-consuming and error-prone step. As a result, most published AI models remain unused in practice, despite strong performance in controlled studies.
This is exactly the gap MHub.ai is designed to fill. It unifies the installation and execution process and requires no manual conversion. It also has a shared ecosystem for benchmarking and improvement, and allows built-in reproducibility checks.
The core pitch of MHub.ai: System Design and Architectural Overview
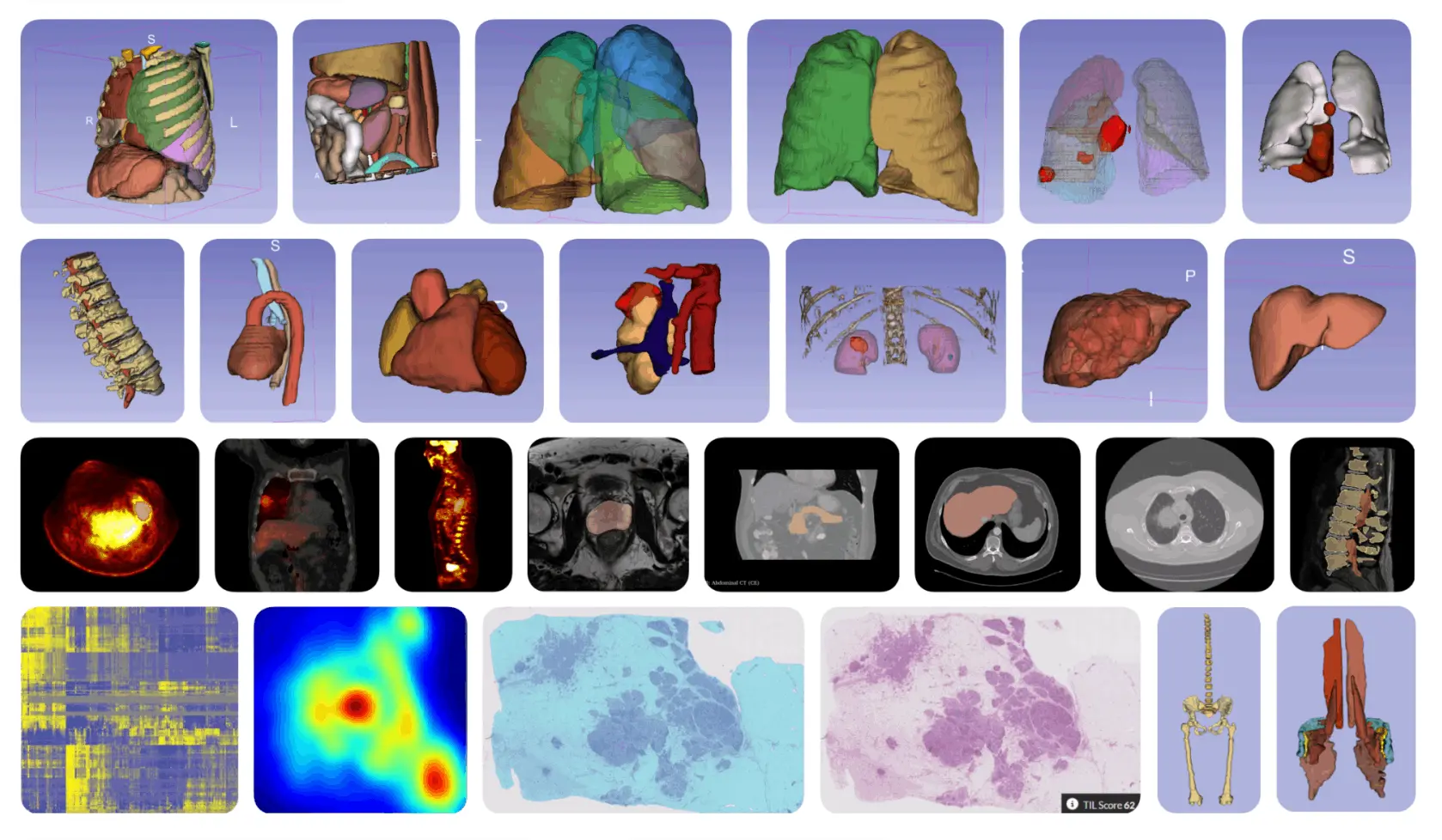
MHub.ai is built as a modular platform with three main components, the first of which is a containerized environment, in which each AI model is wrapped inside a container (similar to Docker) and built on top of the MHub.ai base image. This makes its installation and execution consistent across different systems.
The second major architectural component of MHub.ai is its Unified Application Interface, which provides users with a common way to interact with all AI models in the platform, regardless of their original design; no matter which model is used, users interact with it through the same interface.
This is the most powerful and useful aspect of MHub.ai, as it can directly accept DICOM format as the input and outputs are provided in standardized representations per output type, for example, DICOM-SEG for segmentation, JSON/CSV for tabular data such as predictions, etc.
The third major component that makes MHub.ai stand out from other models is the integration of reproducibility tests to ensure every model in the repository works as intended and produces verifiable results.
Here, each model comes with real-world sample data stored in Zenodo (CERN, 2013), along with the reference results. The framework automatically loads the sample input data and runs the model to generate outputs, which are then compared against the reference outputs. The system then produces both human and machine-readable reports that highlight differences (if any), such as missing or extra files, dice score, or key values.
Case Study: Comparison of Lung Segmentation Models
Authors use lung sedimentation as a proof-of-concept to show the platform’s practical value, as it’s a common and clinically important task in radiology, and multiple groups have already developed models for it.
MHub.ai currently hosts three independently developed AI models for lung segmentation:
- TotalSegmentator
- LungLobes
- LungMask
The authors tested the models on 422 chest CT scans from the NSCLC-radiomics dataset, using Dice Similarity Coefficient (DSC), which is a standard measure of overlap between predicted vs expert annotation; higher DSC meant better segmentation. Out of these, 303 patients had expert-provided left ring lung annotations, which served as the ground truth for comparison.
The models were run side-by-side on DICOM with identical execution codes, so the investigators could directly compare their performance. The case study shows that all models accept DICOM images directly and produce standardized outputs. Each model is paired with reference datasets and reproducibility tests, so results can be validated and compared consistently. Not just this, but the outputs, evaluation metrics, and even generated segmentations are made publicly available through interactive dashboards, allowing others to reproduce the analysis.
All three models achieved high Dice scores, with no clinical differences between male and female patients, suggesting robustness across sex-stratified analyses.
Conclusion
MHub.ai represents more than just a platform—it’s a vision for the future of medical imaging AI. With planned expansions in container runtime support, automated benchmarking, and community-driven contributions, MHub.ai is poised to become an even more powerful ecosystem for researchers and clinicians alike. As these developments unfold, we move closer to a future where AI model validation is truly accessible, reproducible, and scalable across the medical imaging community.
Article Source: Reference Paper | Code Availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.