Cancer, a simple name for a complicated group of diseases, has long perplexed scientists and healthcare professionals alike. Despite years of research and medical advances, individualized therapy remains elusive. However, a recent study using multi-omics data is bringing us closer to understanding cancer’s weaknesses and maybe opening up new therapeutic options. By integrating diverse datasets across 1,768 cancer cell lines representing a spectrum of tumor types, the researchers created a comprehensive map of dependency markers – a treasure trove of potential targets for therapy. This map isn’t a one-size-fits-all blueprint, though. The beauty lies in its ability to reveal pan-cancer markers, common weaknesses shared across different cancer types, as well as tissue-specific markers, and unique vulnerabilities specific to certain tumors.
Introduction
Cancer, a disease as diverse yet so deadly, has long provided a tough challenge to scientists and clinicians. While therapeutic choices have progressed, the elusive ideal of genuinely customized therapy remains out of reach. But a recent study, armed with the powerful lens of multi-omics data, is getting us closer than ever to understanding cancer’s vulnerabilities and leveraging them for therapeutic advantage. This research dives deep into the domain of dependence markers, tiny molecular flags that identify genes critical for a cancer cell’s survival. Think of them as Achilles’ heels – hidden weak areas that, when targeted, could bring down the seemingly unbeatable cancer monster. The study carefully examines four main sources of information: mutation, gene expression, copy number, and pathway activities.
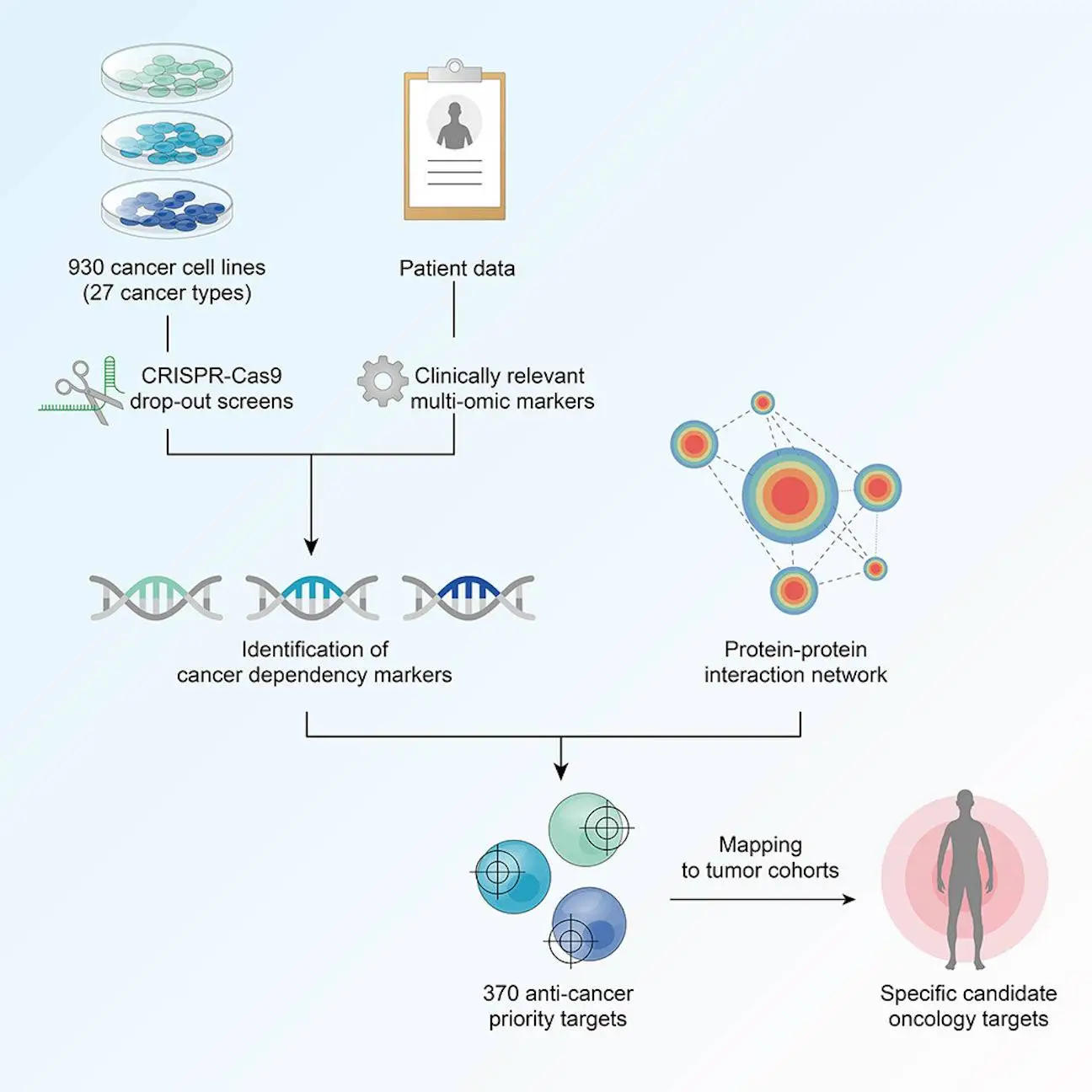
Genetic screens are effective for determining gene function. Large-scale loss-of-function (LoF) screens in hundreds of cancer cell lines with RNAi1 and, more recently, CRISPR2 have revealed fitness or critical genes required for survival across diverse cancer cell types. The systematic study of LoF screens provides insight into the qualities that determine cellular fitness, such as oncogene addictions and paralog associations and genes that display co-essentiality to infer gene function and pathway activities. The researchers offered a comprehensive review of 930 CRISPR-Cas9 screenings done on immortalized human cancer cell lines from many research, identifying and selecting particular gene dependencies for 27 cancer types. Unlike earlier approaches, their study contained:
- Clinically relevant transcriptional patterns
- Metabolic and proteomic data
- A carefully chosen set of cancer driver genes and variations
Furthermore, they used protein-protein interaction networks to connect gene dependencies to molecular indicators and a data-driven strategy to identify particular prospective oncology targets.
A genome-wide survey of gene dependencies in 27 cancer types
The researchers discovered 1,121 pan-cancer core essential genes and an average of 569 genes that were required by all cell lines from specific tissue-identified tissue-specific core essential genes. They discovered that the number of these genes was positively linked with the number of screened cell lines. Discovering three signatures that explained the variations in dependency profiles (excluding core fitness genes): metabolism, apoptosis, and epithelial-mesenchymal transition (EMT).
Curation of clinically relevant multi-omic molecular markers for cancer dependency mapping
The researchers used the Score2 dataset to create a multi-omic assessment of cell lines that included somatic mutation, gene copy quantity and expression, proteomics, transcriptional subtypes, and metabolic profiles. Creating an annotation system to identify possible somatic variants and filter possibly confounding passenger events in 783 cancer driver genes, accounting for somatic mutations, they merged the copy number and gene expression patterns from the Sanger CMP and Broad CCLE datasets.
Comprehensive functional classification of cancer dependency markers
The researchers employed linear regression to find correlations between markers and reliance in the top 500 genes with a selective fitness profile. Found 938 dependency-marker associations (DMAs) for 137 distinct gene dependencies, with an average of two DMAs per dependency, and confirmed that the Score2 dataset was sufficiently powered to detect DMAs in multiple omic datasets.
They developed an upgraded cancer dependency categorization method based on DMAs that takes into consideration new multi-omics markers and their relationship to cancer-associated gain or loss of function genomic events.
Protein-protein interaction (PPI) network analysis reveals functional links between dependencies and markers
PPI networks can illustrate the functional interactions between proteins. To infer functional linkages, researchers projected dependency-marker associations (DMAs) onto a PPI network. Of all DMAs, 34.3% showed a substantial network class, indicating a working link. DMAs with significant functional evidence in the PPI network were statistically more robust.
Second-generation anti-cancer target prioritization framework
Researchers created a methodology to prioritize possible anti-cancer targets based on three criteria:
- Fitness score (impact on cellular fitness following gene deletion)
- DMA score (existence of a dependence marker).
- PPI network score (evidence connecting dependency and marker)
Cancer-type-specific priority targets
The 302 prioritized targets were cancer-type specific. Some prioritized targets had metabolic and proteomic indicators. Marker combinations were found (for example, WRN as a target in colorectal carcinoma linked with RPL22 mutation and other indicators).
Dual markers and composite markers
Some dependencies contained dual markers (e.g., WRN, DNM2, ITGAX). Sixty-five targets have composite markers, providing context for prioritizing. For example, SOX2 in squamous cell lung cancer with loss of TNF-α signaling and elevated MYCL expression.
Validation of priority targets
An orthogonal arrayed screen was used to validate WRN and BRCA2 as priority targets in colorectal cancer with MSI markers.
Key Findings
- Score2 analysis revealed more dependencies than prior efforts, with an emphasis on tumor heterogeneity and significant functional linkages.
- The majority of dependencies were addiction-related (GoF markers), pointing to possible explanations such as increasing cellular reliance or context-specific LoF events.
- PPI network research provides more evidence for functional relationships between markers and dependents.
- Score2 ranked possible targets based on linked markers, enriching for likely causal markers and facilitating multi-omics data integration.
- A candidate target with a DNA biomarker is found in approximately 30% of patients, possibly increasing the number of patients eligible for targeted therapy.
- Patient coverage varies between cancer types, emphasizing the need for novel techniques and the possibility of multi-omics biomarker assays.
Methods
Based on COSMIC20 and IntoGen19 annotations, a proprietary program assessed somatic mutations, filtered germline artifacts, and found driver genes. Binary mutation matrices were created for each of the 783 driver genes. Additional omics data, such as gene expression, copy number, and pathway activity, were analyzed and batch-corrected to ensure consistent analysis. Researchers used linear regression with confounding factor adjustment to link each omic marker to gene dependencies, producing a full DMA matrix. The steps they used are as follows:
- Data Collection and Preprocessing
- Core and Common Essential Gene Identification
- Clustering of Genes and Cell Lines
- Multi-Omic Annotation of Cell Lines in the Study
- Dependency Marker Associations
- Discretization of Continuous Biomarkers
Limitations and Future Prospects
This is the most complete cancer dependence analysis to date, demonstrating the significance of multi-omic data as well as the role of addiction in cancer. The data-driven method combines many data sources to identify viable cancer targets and enhance customized treatment based on tumor features. But as nothing is perfect, it too has some limitations, and they are as follows:
- More cancer models could be included in the analysis, and the limitations of CRISPR screens and cell lines could be addressed.
- Targets should not be disqualified due to a lack of efficacy proof; instead, discovered targets should be considered for future exploration.
- More research is needed to better understand the dependencies caused by non-canonical mutational events and the possibility of combination therapy.
But the journey does not stop here. While this discovery represents a significant milestone, many challenges remain. Additional validation studies in more diverse patient populations are required to confirm the therapeutic usefulness of these indicators. Furthermore, converting these discoveries into successful medications and treatment regimens would necessitate ongoing collaboration among researchers, doctors, and pharmaceutical corporations.
Conclusion
Cancer may have kept its vulnerabilities hidden for years, but a recent multi-omics study has shattered the illusion. Researchers generated a thorough map of dependency indicators – cancer’s hidden Achilles’ heels – by evaluating DNA, gene activity, protein networks, and other data across 1,768 cell lines.
This map identifies both pan-cancer flaws and tumor-specific vulnerabilities, paving the way for tailored treatments. The most promising markers were found through rigorous selection, and network analysis increased their potential as targeted therapeutics. Validation verified these predictions, and patient data indicate that these Achilles’ heels are ubiquitous.
We are witnessing the beginning of a new era in cancer therapy, one in which accuracy and personalization take center stage. This study’s thorough and multifaceted methodology demonstrates the power of scientific collaboration and creativity. It provides a glimpse into a future in which cancer is no longer a single adversary but rather a complicated dance of weaknesses to be understood and exploited. So, let us celebrate this milestone, recognize the researchers’ enormous efforts, and look forward to the next chapter in this riveting scientific drama. Each step ahead brings us closer to a world in which cancer is not only treated but defeated, one tailored Achilles heel at a time.
Article Source: Reference Paper | Reference Article
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.