IntelliGenes is a new machine learning pipeline developed by researchers at The State University of New Jersey that integrates multi-genomics, clinical, and demographic data to discover biomarkers for disease prediction with high accuracy. It implements Intelligent Gene (I-Gene) scoring to assess the significance of biomarkers and create individual profiles to understand disease prognosis. IntelliGenes is an easy-to-use, cross-platform application that enables personalized disease detection and intervention using its innovative multi-genomics models. This pipeline represents a significant advance in the use of artificial intelligence for precision medicine and biomarker discovery.
The advent of high-throughput sequencing technologies has enabled an unprecedented understanding of the genomic basis of human health and disease. Whole genome and transcriptome sequencing can provide a comprehensive readout of an individual’s genetic blueprint and patterns of gene activity. When integrated with clinical data, these multifunctional signatures have great potential to reveal the complex genomic architecture of disease, identify novel biomarkers, and provide more accurate risk prediction and stratification. However, realizing this potential remains a challenge. Traditional statistical and bioinformatics methods often struggle to capture high-dimensional relationships in heterogeneous multiomics and clinical datasets. This is where artificial intelligence (AI) and machine learning (ML) are poised to change the game.
A new study published in the journal Bioinformatics conducted a systematic evaluation of ML techniques for multi-genomic analysis. The findings highlighted two algorithms – support vector machines (SVMs) and random forests (RFs) – as particularly well-suited for handling diverse genomic data types and making accurate predictions. Building on these insights, the researchers have now developed IntelliGenes, an innovative ML pipeline optimized for multi-genomic integration and biomarker discovery.
Introducing the IntelliGenes Platform
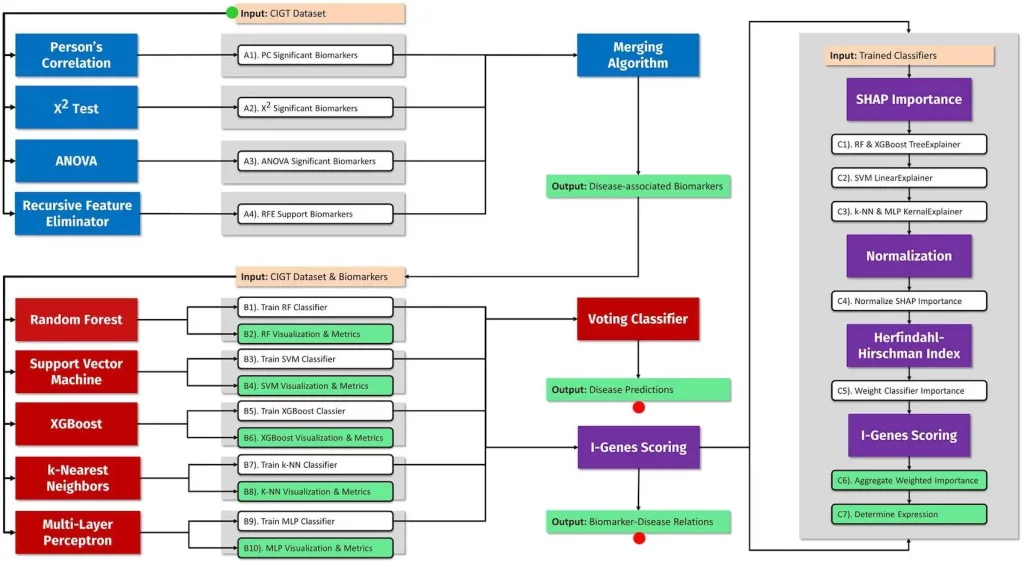
At its core, IntelliGenes employs an ensemble approach, strategically combining multiple ML models with classical statistical methods to extract maximally useful information from multi-genomic profiles. The pipeline takes as input RNA sequencing and clinical and demographic data for a patient cohort. An initial feature selection stage uses statistical tests like ANOVA to identify subsets of genes associated with disease phenotypes. These candidate biomarkers are then passed to an ensemble of RF, SVM, XGBoost, and other advanced classifiers to build high-performance predictive models. Each classifier assigns importance scores to the features, which are aggregated, weighted, and normalized to derive an overall “I-Gene” score representing a gene’s significance in disease prediction.
I-Gene scores delineate a comprehensive footprint of a gene’s predictive capacity, including the directionality of its expression changes between disease and normal states. Researchers can examine I-Gene profiles for individuals to gain a deeper understanding of the genomic factors driving prediction outcomes in a personalized manner. Beyond yielding individual-level insights, when applied across cohorts, I-Gene scoring facilitates the discovery of novel diagnostic and prognostic disease biomarkers.
The researchers demonstrated IntelliGenes’ capabilities on cardiovascular disease datasets, where it achieves up to 96% accuracy in patient stratification. The pipeline uncovers known markers of cardiac phenotypes as well as potential novel transcriptomic biomarkers warranting further functional investigation. Importantly, IntelliGenes is designed for user-friendliness and accessibility. Its modular architecture enables flexible swapping of components to meet diverse experimental needs. The software comes packaged with an intuitive graphical interface, automated visualization, and detailed documentation to guide users.
Applications of IntelliGenes and Future Directions
The research community needs more holistic, multifaceted approaches to unravel the complex genomic architectures of common, complex disorders. Methods that can bridge statistical, biological, and clinical perspectives will be key. The IntelliGenes platform represents an important step in this direction – equipping biomedical researchers with accessible tools to harness multi-genomic data and the power of AI for precision biomarker discovery.
In the future, the research team aims to focus on enhancing IntelliGenes’ capabilities, particularly exploring the integration of genetic variants, epigenomics, and longitudinal information to enable even more accurate disease prediction, subtyping, and therapeutic targeting. A comprehensive 360-degree multi-genomic view of health and disease will be created by incorporating diverse data modalities. This will shed light on the interconnected genomic and environmental influences that drive pathogenesis and progression.
On the computational side, the researchers are actively expanding the library of ML techniques within the IntelliGenes framework. State-of-the-art deep learning architectures like graph neural networks and autoencoders show immense promise for multi-genomic feature extraction and biomarker prioritization. Unsupervised techniques like topological data analysis are also being pursued to enable entirely data-driven stratification of patient cohorts based on heterogeneous molecular profiles.
To enhance accessibility and real-world utility, IntelliGenes will be packaged into a user-friendly web application and low-code environment. This will lower the barriers to entry for biomedical researchers and healthcare teams looking to harness the power of multi-genomic AI. Features like automated data preprocessing, visualization dashboards, and easy sharing of I-Gene biomarker profiles will democratize precision medicine approaches.
Ultimately, IntelliGenes is envisioned to serve as an open platform for the community to build and benchmark novel techniques for integrated multi-genomic modeling and prediction. By fostering collaborative development and putting sophisticated tools into the hands of domain experts, the researchers hope to unravel the complexities of disease and usher in the next generation of AI-driven personalized medicine.
Opportunities and Challenges with Multi-genomic Data Science
The IntelliGenes platform highlights the tremendous opportunities – as well as open challenges – around developing robust, integrative methods for multi-genomic data science. While next-generation profiling technologies continue yielding ever-growing mountains of molecular data, realizing biomedical and clinical value from this wealth of information remains non-trivial. Some key issues that must be tackled include:
- Dealing with high dimensionality and sparsity across diverse datasets: Important signals related to disease pathobiology are often diffuse across thousands of molecular variables. Teasing out these delicate multivariate relationships requires sophisticated feature selection, engineering, and dimensionality reduction techniques.
- Accounting for patient heterogeneity: Disease cohorts can exhibit substantial inter-individual molecular variation. Detecting shared patterns against this backdrop of variability is highly challenging. Adaptive and personalized modeling approaches are needed.
- Limitations of available patient cohort sizes: Assembling datasets with deep multi-genomic profiling at a large scale remains cost and time-prohibitive. This hampers training of complex ML models and biomarker discovery. Data augmentation and transfer learning are avenues to improve data efficiency.
- Interpretability and explainability of model predictions: With AI black-box techniques permeating biomedicine, it is crucial to develop ways of explaining model outputs and extracting biological insights. Approaches like I-Gene signature profiling exemplify steps in the right direction.
- Data integration challenges: Combining heterogeneous data types (sequences, expression profiles, clinical variables) requires extensive multi-modal feature engineering and the development of appropriate similarity measures between datatypes.
- Lack of robust validation of findings: Rigorously validating candidate multi-genomic biomarkers and models on independent datasets is essential to avoid overfitting and false discoveries. This requires expanded patient cohorts and multicenter trials.
While these represent significant challenges, the upside potential for patients is tremendous. Comprehensive multi-genomic characterization coupled with cutting-edge computational methods promises to deliver profoundly better diagnostics, risk predictors, therapies, and a deeper understanding of mechanisms underlying health and disease. The IntelliGenes platform highlights that practical, accessible tools for integrated multi-omic modeling can become a reality. More innovative methods will likely emerge through cross-disciplinary collaboration between biomedical experts, data scientists, and clinicians.
Conclusion
The advent of high-throughput multi-genomics analysis opens up unprecedented opportunities to decipher the genome’s impact on human disease. However, realizing the translational potential of these advanced molecular data requires going beyond traditional bioinformatics approaches. The IntelliGenes platform demonstrates the ability to strategically apply machine learning to integrate disparate sequence, clinical, and demographic information to enable biomarker discovery and personalized risk prediction.
As IntelliGenes continues to improve to address emerging challenges in multi-genomics data science, researchers expect AI-powered comprehensive analysis to be integrated into everything from basic research to the clinic. Computational extraction of key genomic drivers of disease hidden in complex, heterogeneous analytical data will be key to tailoring more accurate diagnostics, prognoses, and treatment targets to each patient’s unique biological basis. IntelliGenes is a major step towards realizing the vision of predictive, personalized medicine powered by artificial intelligence.
Article Source: Reference Paper | IntelliGenes source code is available on GitHub and Code Ocean
Follow Us!
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.















