In order to establish standardized high-throughput whole-genome sequencing (WGS) for patients with cancer and rare diseases, the UK Government launched the groundbreaking 100,000 Genomes Project within the National Health Service (NHS) in England. This was accomplished through the use of an automated bioinformatics pipeline accredited by the International Organisation for Standardisation. Operating in conjunction with NHS England, Genomics England examined WGS data from 13,880 solid tumors representing 33 different cancer types, fusing genetic information with actual treatment and outcome data in a secure research environment. WGS and longitudinal life course clinical data were linked, enabling the evaluation of treatment results for patients categorized by pangenomic markers. The results of this study show how useful it is to connect genomic and practical clinical data in order to do survival analysis, locate cancer genes that influence prognosis, and deepen our knowledge of how cancer genomics affects patient outcomes.
Goal of the 100,000 Genomes Project
Using an automated bioinformatics pipeline accredited by the International Organisation for Standardisation, the 100,000 Genomes Project was a ground-breaking UK Government initiative implemented within the National Health Service (NHS) in England with the goal of establishing standardized high-throughput whole-genome sequencing (WGS) for patients with cancer and rare diseases.
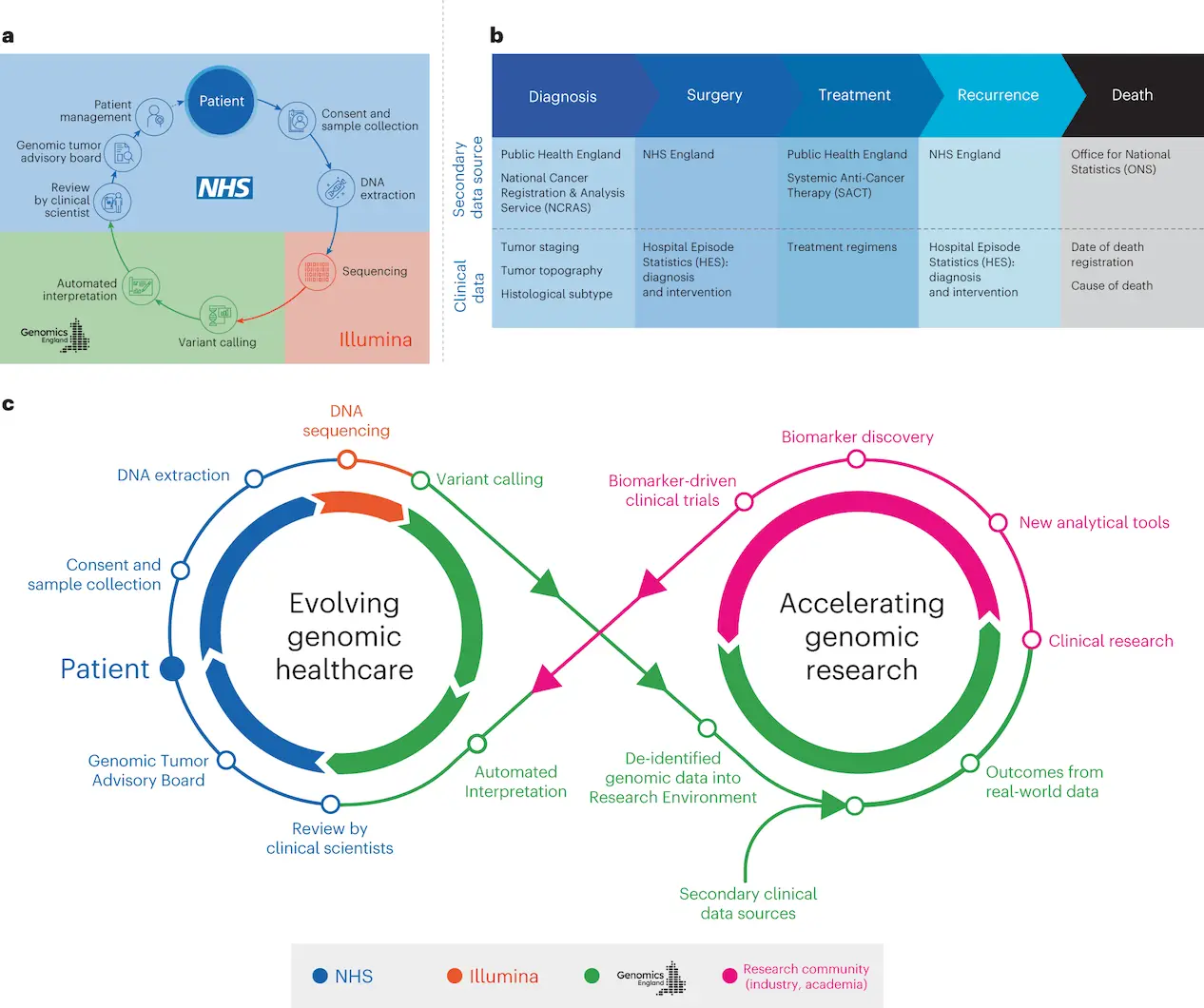
The 100,000 Genomes Project’s Cancer Programme assessed Whole Genome Sequencing’s (WGS) usefulness in the NHS for cancer patients. In a secure study environment, participants gave signed, informed consent for genomic data to be shared with researchers and linked to anonymized health records. The National Genomic Research Library, a nationwide molecular data platform with secure links to long-term public data, was established using the data. This strategy enables genetic research and discoveries to be fed back into genomic healthcare, boosting the understanding of diverse cancer diseases.
WGS-based Clinical Actionability
The Cancer Programme uses a single test termed WGS to identify somatic minor variations, copy number aberrations, and structural variants by combining tumor-normal sequencing. A higher yield of therapeutically meaningful results is possible with this genomic test since it combines pharmacogenomic and germline data. The Cancer Programme provides standardized WGA results to NHS GMC Laboratories, which are subsequently examined by interdisciplinary Genomic Tumor Advisory Boards (GTABs) and clinical scientists for insights that can be put into practice.
Pharmacogenomic analysis of 13,880 whole genomes identified a variety of mutations associated with hereditary cancer risk, such as fusions, CNAs, and minor variations. Germline mutations linked to hereditary cancer risk have been found by the National Genomic Test Directory for Cancer (NGTDC). Although it varied, a significant portion of patients had somatic mutations in genes listed in the NGTDC. For instance, in glioblastoma multiforme, low-grade glioma, skin cutaneous melanoma, head and neck squamous cell carcinoma, colon and rectal adenocarcinoma, and lung adenocarcinoma, more than 50% of tumors carried one or more mutations identified in genes recommended for testing in the NGTDC. Less than 20% of cases in other cancer types, such as pancreatic, prostate, esophageal, and stomach adenocarcinomas, had mutations in genes present in the NGTDC.
In contrast, 20–49% of cases of lung squamous cell carcinoma, ovarian high-grade serous carcinoma, uterine endometrial, sarcoma, mesothelioma, and bladder urothelial carcinoma had clinically relevant mutations. The clinical actionability of mutations is influenced by individual case characteristics and clinical situations. The interpretation plays a pivotal role in ascertaining the suitability of a treatment plan, underscoring the necessity of meticulous deliberation within a GTAB.
Researchers at GTAB have discovered mutations in the NGTDC in a number of cancer forms, including tumor mutational load and HRD. These variations can result in additional GTAB reviews or clinical studies. SNVs in PIK3CA and KRAS, as well as pangenomic indicators like HRD and TMB, could be targeted for clinical trials. NGTDC indications are anticipated to broaden, embracing new genes and biomarkers across a range of cancer types as evidence from biomarker-driven trials increases.
Applications of Research Environment
- Approved researchers get secure access to genetic data and related health data through the Research Environment, a platform developed by Genomics England and NHSE. It has made fundamental research advancements possible, such as the identification of genes that cause cancer, mutational signatures, or modifications to clinical practice brought about by the accessibility of WGS testing.
- To find all clinically significant mutations for a particular disease and their connection to licensed precision medicines in order to maximize cancer care.
- Prognostic and predictive molecular markers can be refined not only by combining various genomic abnormalities but also by utilizing developing technologies to broaden the application of precision oncology and enhance cancer outcomes.
Knowing about GTAB
GTABs play a crucial role in examining eligibility for approved medicines and clinical trials and making sure that actionable data are shared with treating teams and physicians. Clinical interpretation of cancer genomic testing is greatly aided by a well-designed and organized GTAB, which can also help with clinical trial enrollment, offer advice to clinicians, and possibly improve results. This strategy fits nicely with adaptive basket studies like DETERMINE, which was created to assess approved therapies in unapproved indications like the DRUP trial. In addition to ensuring that patients are fully considered for clinical research and trials as a result of this genomic testing, the goal is to enable more equitable and thorough molecular testing within the NHS and to optimize cancer care by identifying all clinically relevant mutations for particular cancer and their relationship to approved precision medicines. Furthermore, the goal is to explore clinical trial options, including the use of repurposed, well-known, and well-characterized drugs.
Limitations in implementing clinical WGS
- Expensive.
- Requirement of operational equipment.
- Less availability of skilled and knowledgeable multi-professional workforce.
More about WGS
- The commissioning of clinical WGS for sarcoma, glioblastoma, ovarian high-grade serous carcinoma, and triple-negative breast malignancies was supported by the evaluation of WGS data. This genomic testing is used to identify many types of mutations, including pangenomic markers, and provides information to guide clinical management.
- A lot of evidence supports the co-occurrence of CNAs, indels, and SNVs. The co-occurrence of CNAs and somatic minor variations affecting cancer genes in the NGTDC was investigated using WGS.
- By connecting the WGS data to clinical data from the longitudinal life course (SACT and ONS), researchers evaluated the results of treatment for individuals categorized based on pangenomic markers.
- WGS data characterizes the clinical genomic landscape of a tumor.
- WGS allows the simultaneous detection of germline and somatic variants.
- Multidisciplinary Molecular Tumor Boards, or GTABs, convene to examine WGS results in order to assess somatic and germline variations, ascertain clinical actionability, and offer therapeutic recommendations.
Conclusion
The 100,000 Genomes Project generated valuable insights by combining whole-genome sequencing data from nearly 14,000 tumor samples with real-world clinical data. The combination of genomic and clinical analysis deepens our understanding of triggering mutations in various cancers, identifies prognostic genomic signatures, and enables assessment of outcomes in genomically stratified patient populations. This groundbreaking dataset demonstrates the ability to link genome-wide profiling with longitudinal clinical data to reveal the genomic landscape of cancer, guide precision oncology, and ultimately improve patient care and outcomes. The project provides a model for large-scale genomic initiatives that aim to use multidimensional data to advance personalized cancer care.
Article Source: Reference Paper | Reference Article
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.
















[…] Integrating Genomic and Clinical Data from the 100,000 Genomes Cancer Program Paves the Way for Prec… […]