Drug discovery, synthetic biology, and enzyme engineering are all dependent on the ability to comprehend and create biomolecules, such as proteins and tiny molecules. Recent advances in Artificial Intelligence (AI) have brought biomolecular research to a new level of precision in biomolecular design and prediction. However, a significant divide still exists between the computational prowess of AI and the intuition of researchers who use natural language to match the intents of humans with molecular intricacy. Large Language Models (LLMs) have demonstrated the ability to interpret human intentions; however, because of various obstacles, such as the need for specialized knowledge, multimodal data integration, and semantic alignment between natural language and biomolecules, their application to biomolecular research is still in its infancy. In order to overcome these constraints, Zhejiang University researchers present InstructBioMol, a revolutionary LLM that aims to provide a thorough, any-to-any alignment of plain language, molecules, and proteins to bridge the gap between science and biomolecules. With the help of this model, researchers may precisely address biological requirements by producing biomolecular outputs that incorporate multimodal biomolecules as input and allow them to express their design aims in natural language.
Introduction
Research in natural sciences requires an understanding of the ability to design biomolecules. The exact manipulation of biomolecules—such as proteins and tiny molecules—is crucial to the growth of synthetic biology, enzyme engineering, and drug discovery since these molecules are vital to biological processes. Recent advances in Artificial Intelligence (AI) have revolutionized these fields of study. Biomolecular structure prediction has undergone a revolution thanks to programs like AlphaFold3 and RoseTTAFold All-Atom, which provide previously unheard-of speed and precision. Notwithstanding these developments, a critical problem still needs to be solved: how to successfully comprehend biomolecules using natural language and build them with human purposes in mind. This creates a disconnect between the computational capabilities of AI and the necessity for academics to use them to solve practical issues. Imagine a scientist is assigned to create a novel medication targeting a protein implicated in a complicated illness. In the past, this method involved sorting through enormous volumes of biomolecular data, deciphering chemical and biological linkages, and iteratively creating molecules with desired characteristics by trial and error. While AI has improved this workflow in many ways, present solutions frequently fail to match molecular complexity with clear goals driven by humans and expressed in normal language.
Looking into LLM
Large Language Models (LLMs) have proven to be remarkably adept at comprehending human purpose and producing safe, helpful replies that resemble those of a human.
Due to their many learnable characteristics and great alignment with human preferences, large language models, or LLMs, have a remarkable ability to comprehend human intentions and produce responses similar to those of humans. However, there are obstacles to overcome before using LLMs for biomolecular research may be undertaken. These include the requirement for specialized expertise, the lexical and semantic differences between natural language and biomolecules, and the necessity of aligning human intention in biomolecular tasks. In order to help LLMs achieve greater comprehension and more accurate performance of particular jobs, instruction is essential to this alignment process. For difficult activities like building compounds for target proteins or generating enzymes for substrates, mastering the alignment of chemicals, proteins, and natural language is crucial.
Understanding InstructBioMol
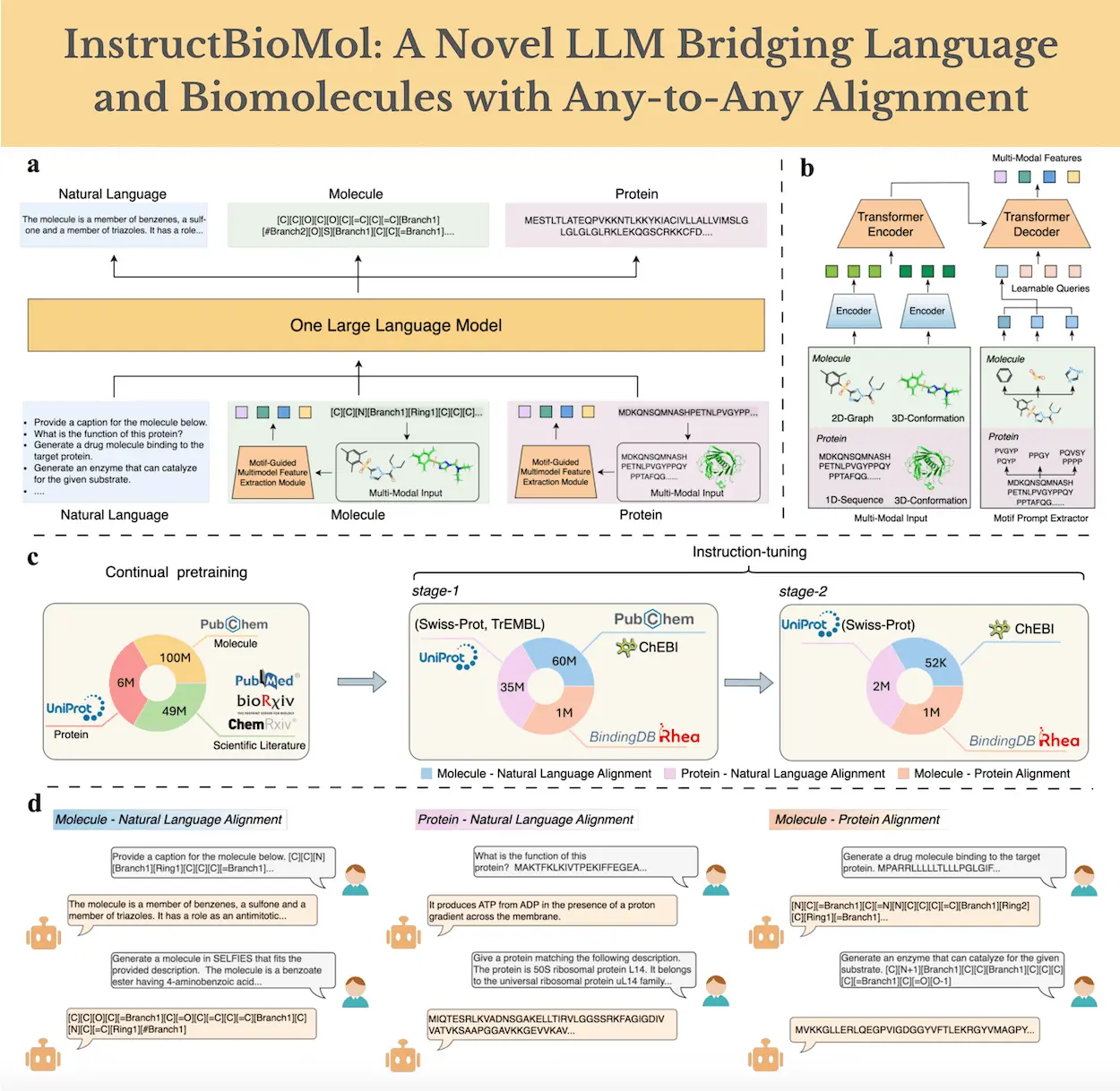
Natural language and biomolecules are integrated by InstructBioMol, a large language model that achieves any-to-any pairwise alignment between natural language, molecules, and proteins. It interprets and designs biomolecules in accordance with human intention by utilizing a carefully selected dataset of instruction settings at the hundred-million scale. Additionally, the model suggests a motif-guided multimodal feature extraction module that can extract a variety of features, including 1D sequence and 3D geometric properties of proteins, as well as 2D topological and 3D geometric aspects of molecules. As a virtual research assistant, InstructBioMol helps scientists with their biomolecular investigations and discoveries. It is particularly good at using natural language processing to investigate biomolecules, providing answers to queries about the activities of proteins, and creating new compounds from written descriptions.
The architecture of InstructBioMol: An Overview
The Biomolecular Vocabulary-expanded Language Model and the Motif-Guided Multimodal Feature Extraction Module make up the two halves of InstructBioMol’s architecture. The former is made to extract biomolecules’ multimodal features, while the latter manages the integrated processing of textual natural language, protein, and molecular data in addition to the multimodal features that have been extracted. In particular, researchers utilize lightweight frozen pre-trained encoders in the Motif-Guided Multimodal Feature Extraction Module to extract features from each modality independently. Scientists then use the biological knowledge incorporated into motifs to direct the fusion of these multimodal features. Researchers expand the vocabulary to include molecules and proteins in the Biomolecular Vocabulary-expanded Language Model and standardize the input format for their multimodal properties in order to reduce the possibility of interference between data from different domains.
Conclusion
The goal of the multimodal language model InstructBioMol is to comprehend and create biomolecules in accordance with human directives. It extracts multimodal features from biomolecules and incorporates them into the language model using a motif-guided multimodal feature extraction module. In order to accomplish any-to-any alignment between natural language, molecules, and proteins, the model employs a continuous pretraining followed by an instruction-tuning technique. This method improves quality without increasing data size. Drug-like compounds for target proteins or enzyme catalysts for reaction substrates can be designed by InstructBioMol. Because of this, it serves as a useful research collaborator that provides insightful analysis and motivation to scientists working on drug and enzyme creation. The model’s ability to achieve any-to-any alignment on a variety of tasks indicates that it could be a useful research copilot.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.