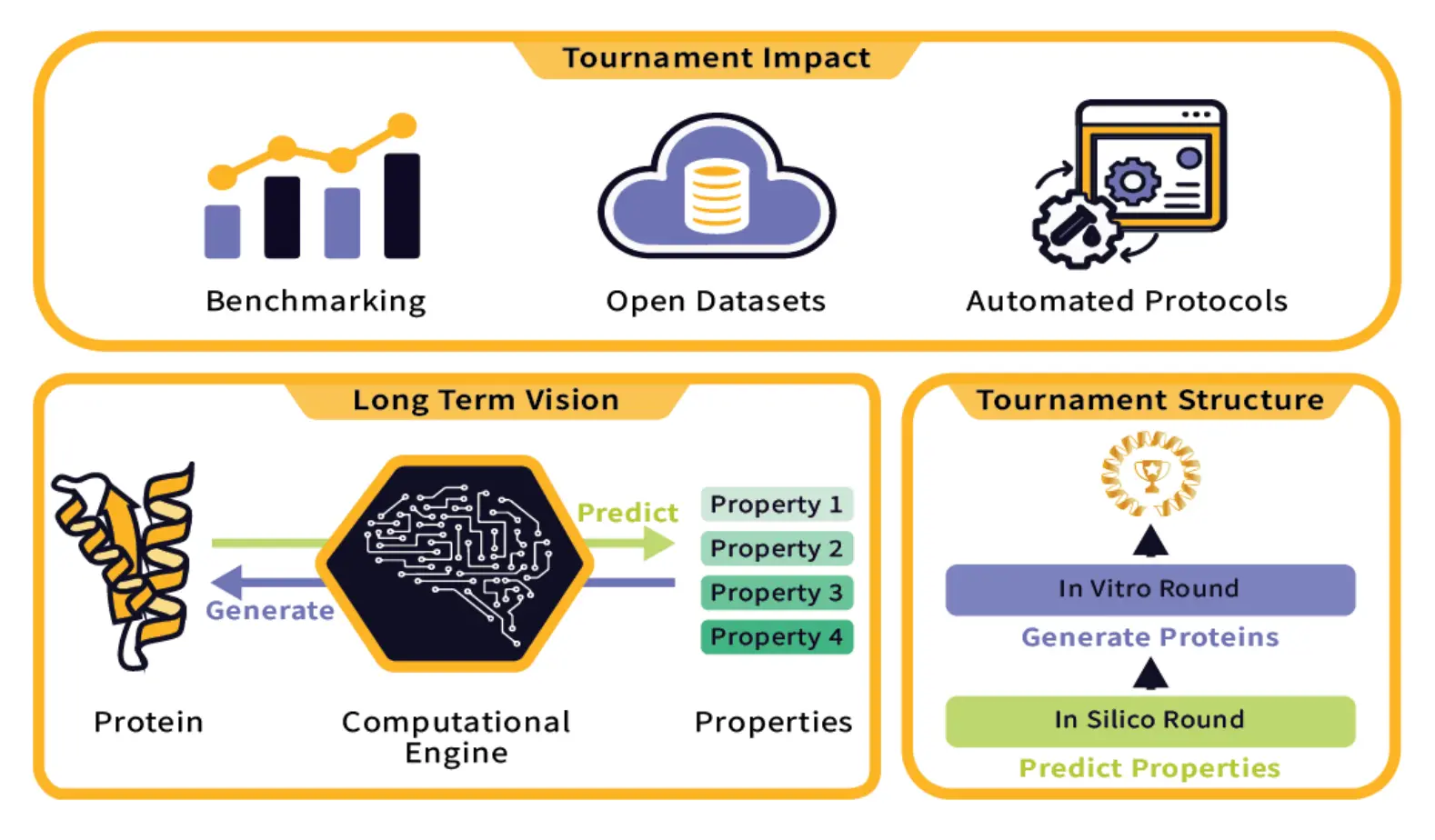
Protein engineering heavily relies on generating sequences using computational models. Limitations such as lack of benchmarking opportunities, scarcity of large protein function datasets, and limited access to experimental protein characterization slow down the progress in this field. To overcome these challenges, the Protein Engineering Tournament was established to prioritize computational methods in protein engineering. The competition was organized by a collaboration of top institutions, such as Columbia University, Medium Biosciences, Align to Innovate, the Department of Systems Biology at Harvard Medical School, and many others.
The Protein Engineering Grand Challenge
The ultimate vision for protein engineering is to have computational models that can essentially predict and design protein sequences for any function with high accuracy. The achievement of the latter requires not only good algorithms but also rich datasets with robust experimental validation. Again, fragmented efforts in many individual labs that deal with proprietary datasets using methods without standardized benchmarks have traditionally hampered this work. It is further compounded by not only a limitation in collaboration but also a lack of open sharing of data, which makes assessing true progress in the field and real identification of the most promising approaches an almost insurmountable task. Concerning these, this tournament was established as a fully remote competition, which consisted of an in-silico round (to predict biophysical properties from protein sequences) and an in-vitro round (to design novel protein sequences).
Entrants came from academia, industry, and research, but they all had only one wish in common: to reach the farthest point of what can be achieved with protein engineering.
Structure and Aims
Previously, science competitions such as Critical Assessment of Structure Protein (CASP) and Critical Assessment of Computational Hit-finding Experiments (CACHE) focused on just benchmarking models. While designing the Protein Engineering Tournament, several important objectives had to be kept in view. First and foremost, it provided a test bed for benchmarking different computational approaches toward protein modeling (similar to CASP and CACHE). This was done by formulating a set of challenges wherein participants were required to predict protein sequences meeting certain functional criteria. The predictions go through validation against experimental data to get a clear, objective measure of each of their performances.
It also aimed to encourage the creation of novel datasets and benchmarks that the research community at large could make use of in the future. Indeed, sharing data and results openly would, therefore, also enable the organizers to create an interesting asset for the entire research community.
Finally, the tournament was designed to improve collaboration and knowledge transfer among participants. Workshops, seminars, and online discussions gave the participants an opportunity to learn from each other, share their insights, and fine-tune their approaches. That is one of the beautiful features of the tournament: it speaks to the belief that grand challenges in protein engineering can be tackled only if we do it collectively.
Results and Impact
All teams engineered variants with improved activities, but TUM Rostlab came in first by generating the top score of an individual variant and the highest median of all qualifying variants according to the combined criteria for expression and thermostability. MediumBio and Marks Lab finished second and third, respectively.
This contest proved that open science and collaboration have great power to move protein engineering as a field. By bringing many players into the game and with a common platform for testing, the tournament could tease out the most promising approaches in protein modeling and design.
The competition also made experimental validation more valuable in protein engineering, which has largely been ignored. Computational models drive the prediction of protein sequences, but such predictions need empirical data against which they can be tested for their reliability and accuracy. The one during the tournament put long-needed reality checks on so many of the models in a way that led them to better, more accurate algorithms.
Besides, all data and associated benchmarks developed through the tournament are in the public domain and are shared with the wider research community. This open-access model is expected to be an enduring strength of the field and will have practical benefits for researchers at the forefront of driving accelerated discovery in protein engineering.
Conclusion
This is the landmark of study in protein engineering: the Protein Engineering Tournament. By incorporating some of the critical challenges facing the field—lack of standardization of benchmarks, poor access to larger datasets, and the requirement for experimental validation—the tournament set new rigorous and collaborative standards for protein modeling and design. An initiative like this, as the field continues to emerge, will ensure that protein engineering can really come of age, delivering new proteins with functions that could have an enormous impact on medicine, industry, and the environment.
The outcome of the Protein Engineering Tournament attests to the value of open science and the power of collaboration in fostering innovation. Infusing the competition with collaboration and knowledge sharing has advanced not only the field but also set the bar high for most scientific research. Entering the future, we can only be assured that the challenges posed by tomorrow will have to be solved by the teamwork of the global research community to unlock the vast potential that exists within protein engineering.
Article Source: Reference Paper | All team submissions and datasets are available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!