The quest to understand biomolecular interactions is central to advancing drug discovery, molecular biology, and bioengineering. A significant step in this journey is accurately predicting the 3D structures of biomolecular complexes. Researchers from Iambic Therapeutics recently unveiled NeuralPLexer3 (NP3), a groundbreaking model designed to set a new benchmark in structure prediction by leveraging flow-based models. This study aimed to overcome longstanding challenges in accurately modeling biomolecular interactions and confidence assessment.
Why NeuralPLexer3?
Many established techniques in structure prediction fail to detail the molecular features, especially for out-of-equilibrium or complex systems. However, considering the number of experimental structures being published and their increased availability, it became the talk of the day that there is a need for a user-friendly but comprehensive tool that would encompass all insights while growing to something complex. NP3 was created to address this issue by combining cutting-edge neural designs and training methodologies to meet the expansion target.
The Architecture Behind NP3
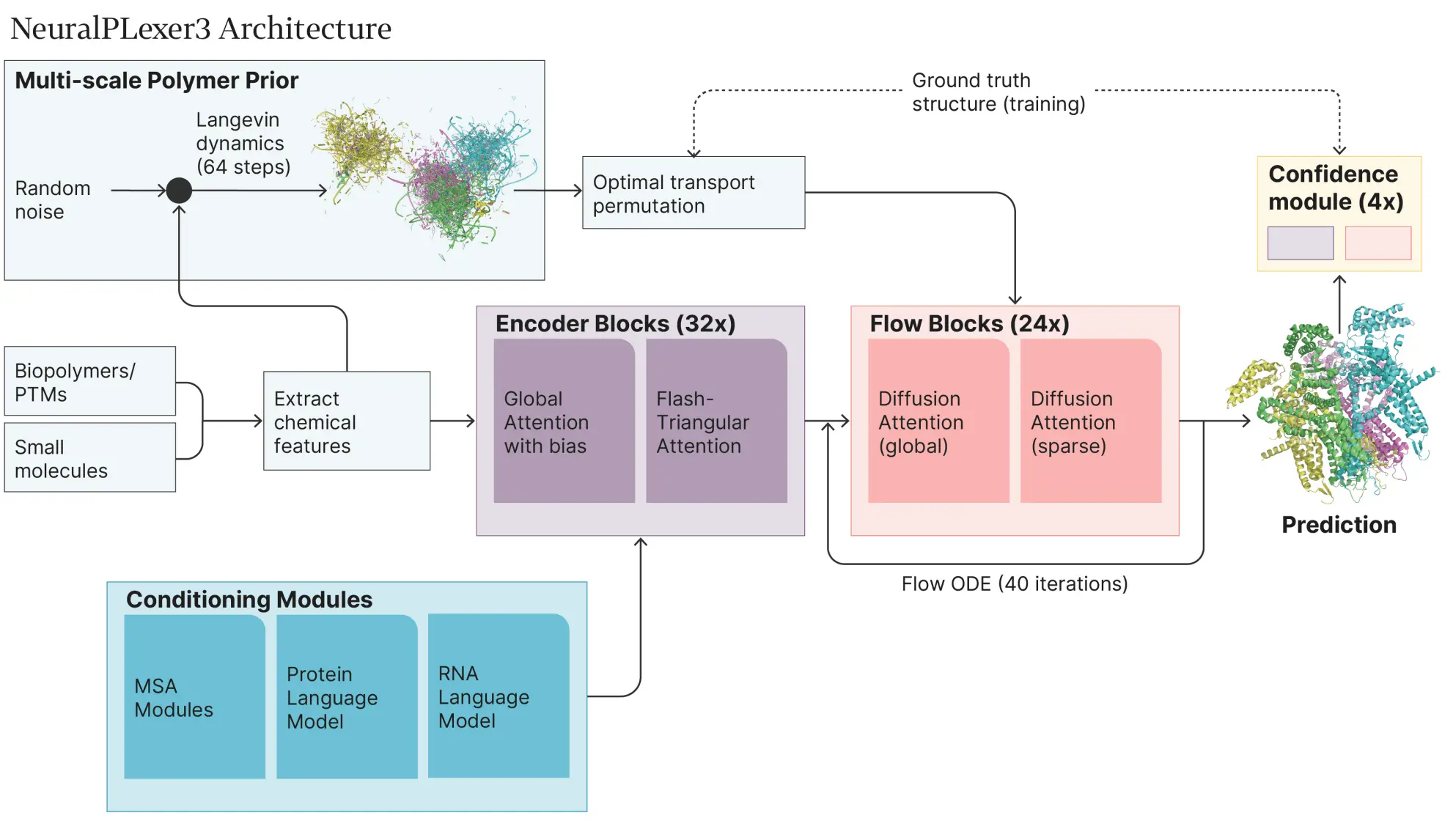
NP3 is designed with an expandable multilayer architecture that includes advanced structural features, the treatment of noise, and confidence scoring. It uses PyTorch FSDP supported with BF16 mixed precision to preserve resources during its bulk training over 64 NVIDIA H100 graphics processing units. For more than twenty-four days, NP3 went through an iterative structure cropping process, during which its capability to resolve longer molecular contexts was gradually enhanced. Most training evolved with smaller crop sizes of 384 anchors or 3072 atoms over a large bulk of time, moving to the latter stages where 1024 anchors or 12800 atoms were much larger contexts.
To address the issues of memory and the resolution of the context that one targets, NP3 employed the Flash-TriangularAttention kernel, which effectively prevents the training GPU from memory overflowing during training large structure crops. In the same model, activation checkpointing would enhance the activity of the model without compromising memory usage.
Confidence Module: Enhancing Prediction Reliability
A unique highlight of NP3 is its Confidence Module, designed to evaluate the reliability of its predictions. This module outputs two metrics of confidence, which are the predicted local distance difference test (pLDDT) for individual atoms and the predicted distance error (pDE) for anchor pairs. The module combines noise sampling and cross-entropy loss optimization against structural error to achieve consistent confidence scoring.
The new training paradigm was based on producing 3D noisy molecular structures, molecular structures prediction based on the NP3 model, and adjustment of confidence scores with respect to true errors, which helped in improving the ability to discriminate different conformations and, more importantly, made it possible for the model to quantitatively judge different predictions of the targets.
Practical Applications and Benchmarking
To test NP3’s skills, the researchers briefly prepared two benchmark datasets: NPBench and ConfBench. The focus of NPBench was on the determination of accuracy regarding the more recent PDB depositions, whereas ConfBench was specifically designed to assess the likelihood of predicting ligand-induced conformational alterations of the model. Each of the sets went through a rigorous filtration process to have high-quality and non-overlapping targets, which provided minimal evaluation requirements.
NP3 outscored the multi-model AlphaFold, which was considered among several models to have performed top-notch accuracy and confidence evaluations. In this match, for instance, in ConfBench, NP3 was able to predict large conformational change with a 47.7% win rate of structures that were highly diverged(scores>0.5), while Alpha Fold managed only a score of 32.2%.
Among the parameters used in assessing NP3 were pocket-aligned ligand RMSD, DockQ scores, generalized RMSD, TM-score, and LDDT scores. These studies gave NP3 an edge in modeling larger and more local structural features.
Conclusion
The launch of NP3 is not only a technical achievement but a facial revolution of the synthesis with biomolecular interactions at an unprecedented level of detail. NP3 overcomes the challenges of the currently available prediction models and adds appropriate confidence scoring, thus creating fresh opportunities for drug discovery, protein engineering, and other biotechnological advances. The ability to reliably predict the configuration of intricate structures may pave the way for the novel treatment and foster insights into primary biological paradigms.
The investigation done in Iambic Therapeutics is a wonderful example of true interdisciplinary teamwork, where computation is put to effective use and engaged in the context of biology. NeuralPLexer3 establishes a new benchmark in predicting structures of biomolecules, which is bound to change the face of isto bioinformatics and structural biology as we know it today.
Article Source: Reference Paper | Reference Article | The NPBench repository is accessible on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.