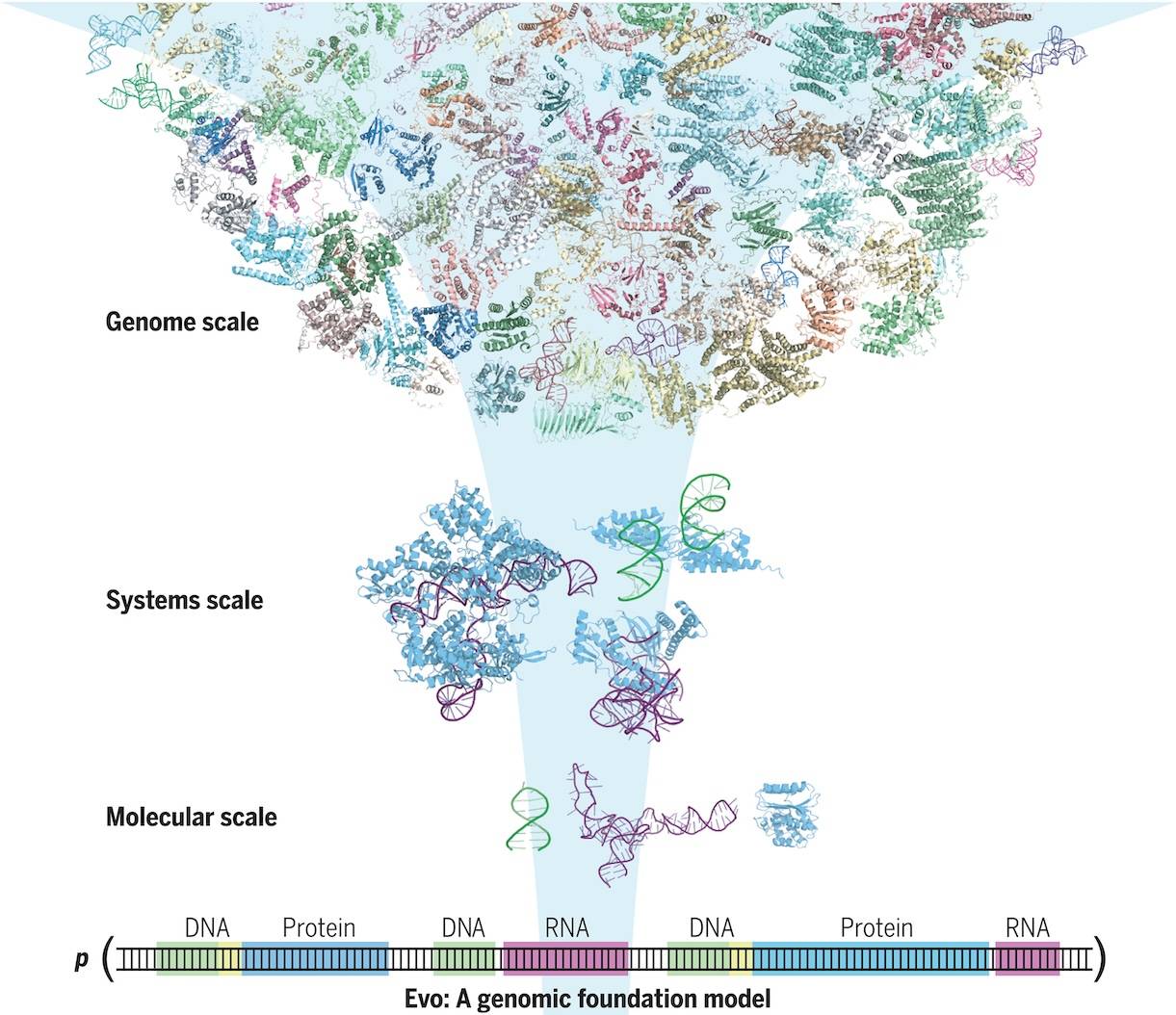
The DNA sequences of all living organisms hold the fundamental instructions for life. Massive datasets of entire genomes, together with machine learning advancements, may allow for the creation of a biological foundation model that accelerates the generative design and mechanistic comprehension of intricate genetic relationships. The researchers from Arc Institute and Stanford University, USA, introduced Evo, a genomic foundation model that allows for prediction and generation tasks at the molecular to genome scale. At single-nucleotide byte resolution, the deep signal processing architecture Evo has been scaled to 7 billion parameters with a context length of 131 kilobases (kb).
Evo, which has been trained on entire prokaryotic genomes, can generalize across the three basic modalities of molecular biology and exceed or compete with the best domain-specific language models in zero-shot function prediction. For the first time, Evo can create whole transposable systems and synthetic CRISPR-Cas molecular complexes, demonstrating its proficiency in multielement creation tasks. Additionally, it can produce coding-rich sequences up to 650 kb in length and predict gene essentiality at nucleotide resolution.
Introduction
The fundamental layer of biological information has been revealed through the systematic mapping of evolutionary variation at the whole-genome scale, made possible by the rapid advancements in DNA sequencing technologies. At the phenotypic level, adaptation and selection for biological function are reflected in evolutionary variation in genomic sequences. A generic biological foundation model that understands the inherent logic of entire genomes may be made possible by combining machine learning algorithms and enormous genomic sequence datasets. The need for more molecular biology research is highlighted by the fact that current efforts concentrate on developing modality-specific models tailored to proteins, regulatory DNA, or RNA.
In generative applications in biology, transformers are essential, especially in intricate biological processes like genetic transposition, CRISPR immunity, and gene regulation. Several interactions between molecules in various modalities are necessary for these activities. Designing more complex biological activities is made possible by DNA models’ ability to learn from vast genomic regions through the integration of data from molecular systems, as well as genome sizes. The evolutionary consequences of sequence variation, such as specific single-nucleotide mutations that change organism function, can be incorporated into these models because they operate at single-nucleotide resolution. Single-nucleotide precision is sacrificed in Transformer-based DNA models, which are limited to small context lengths and employ techniques that aggregate nucleotides into tokens. Transformers could transform DNA model design and allow for more complex biological functions despite these drawbacks.
Diving Deep into Evo
This paper introduces Evo, a genomic foundation model with seven billion parameters that is taught to produce DNA sequences at the whole-genome scale. Evo is built on the StripedHyena architecture, which effectively combines data-controlled convolutional operators with attention to process and recall patterns in lengthy sequences. It employs a context length of 131k tokens. Using a byte-level, single-nucleotide tokenizer, Evo is trained on a 300 billion nucleotide bacterial whole-genome dataset.
Evo is a new paradigm that may be applied to issues involving generation and prediction at the molecular, system, and genome levels. It can predict combinations of prokaryotic promoter-ribosome binding site pairs that result in active gene expression, outperform specialized RNA language models in predicting fitness effects of mutations on noncoding RNAs, and outperform state-of-the-art protein language models in predicting fitness effects of mutations on E. coli proteins. To create synthetic multi-component biological systems, Evo also learns the co-evolutionary relationship between coding and noncoding sequences. Evo is capable of autonomously predicting key genes in bacteria or bacteriophages at the whole-genome level. A credible genomic coding architecture can produce sequences longer than 650 kilobases. The ability to engineer life will accelerate, and a greater comprehension of biology will be made possible by further development of Evo.
Limitations
Despite its impressive potential, Evo, a first-generation DNA foundation model, has certain technological issues. 300B prokaryotic tokens, a tiny subset of the genomic data that is publically accessible, are used to train the model. Determining the functional implications of mutations on human proteins’ fitness is limited. Evo can also exhibit traits that natural language models can, such as sustaining diverse and coherent production over extended periods. Despite producing hundreds of kilobases at genome size, Evo finds it difficult to incorporate important flag genes such as complete tRNA-encoding repertoires. These restrictions are similar to those of natural language models, which have been refined over time through labeled data, prompt engineering, human preference alignment, and larger scale.
Future Directions
Evo provides an extra size and resolution that will open up a wide range of research avenues. The development of systems biology properties may result from its capacity to enhance molecular structure prediction and direct the evolution of multi-gene biological systems. Additionally, Evo might serve as the foundation for a next-generation sequence search method, allowing for relational or semantic metagenomic mining. However, it takes a significant investment of resources in engineering, computation, and safety-related model alignment to integrate eukaryotic genomes into Evo. The ability of Evo to advance biological engineering is demonstrated by the fact that, despite the complexity of these genomes, it extends the reach of biological engineering and design to the size of entire genomes.
Conclusion
The genomic foundation model Evo creates and predicts DNA sequences at the scale of individual molecules, chemical complexes, biological systems, and entire genomes. It has been trained on hundreds of billions of DNA tokens from prokaryotic life’s evolutionary diversity. Single nucleotide resolution language modeling at a context length of 131k is made possible by Evo’s cutting-edge hybrid model architecture, StripedHyena. It performs better at every scale level than baseline architectures, such as the Transformer architecture. With the ability to sample CRISPR-Cas proteins, noncoding guide RNAs, multi-gene transposable systems, and 650 kb sequences, Evo is also a generative model.
Article Source: Reference Paper Abstract | Full Paper bioRxiv | Open-source code and models for Evo are publicly available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.