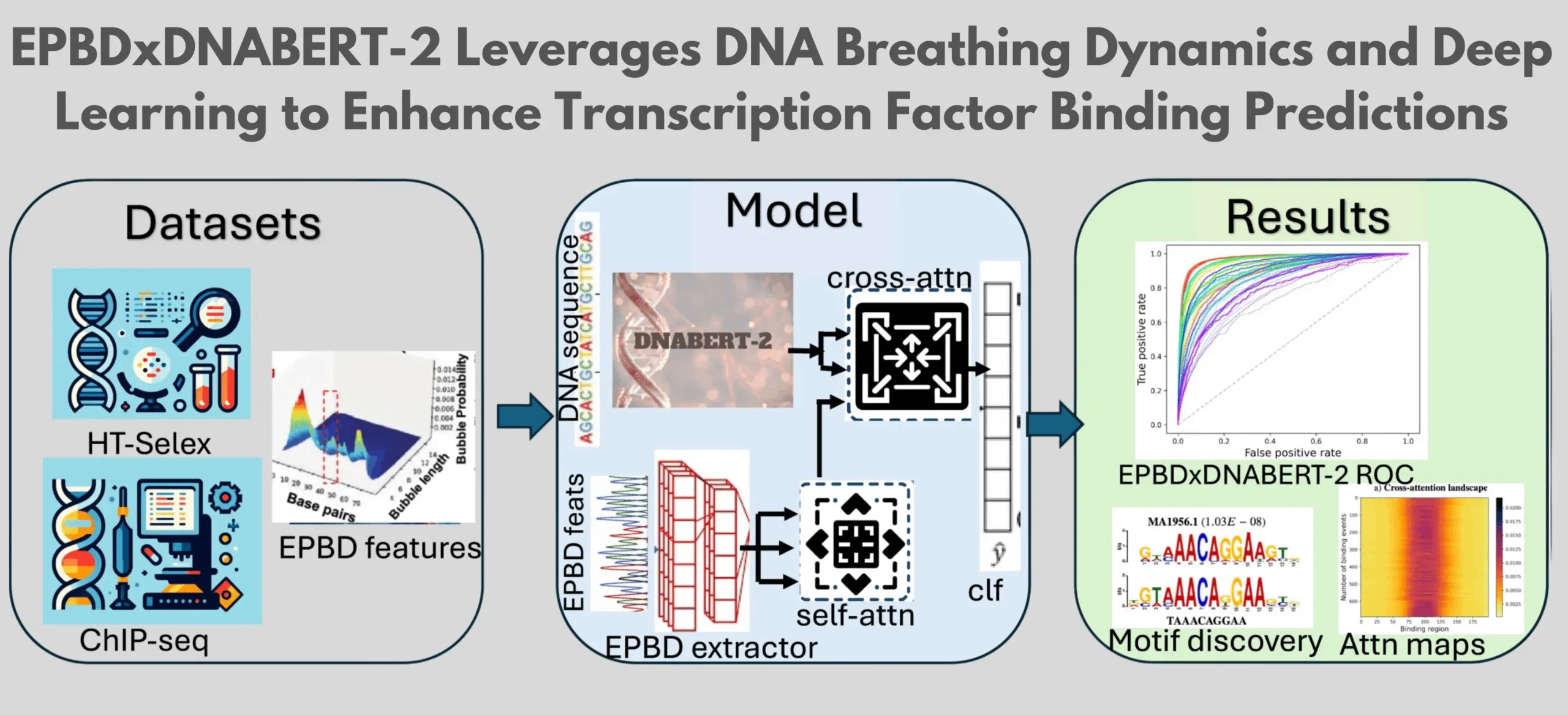
It has been demonstrated that there is a functional correlation between transcriptional activity, thermodynamic stability, DNA breathing, and transcription factor (TF) binds. The researchers created the multi-modal deep learning model EPBDxDNABERT-2, which is based on the Extended Peyrard-Bishop-Dauxois (EPBD) nonlinear DNA dynamics model, to determine the exact link between TF binding and DNA breathing. With an increase in the area under the receiver operating characteristic (AUROC) metric of up to 9.6% when compared to the baseline model, the EPBDxDNABERT-2 model, which was trained using chromatin immunoprecipitation sequencing (ChIP-Seq) data, considerably improves the prediction of over 660 TF-DNA variations. In order to compare it with existing frameworks, the model was further extended to the in vitro high-throughput Systematic Evolution of Ligands by Exponential enrichment (HT-SELEX) dataset, which included 215 TFs from 27 families. The incorporation of DNA breathing characteristics into the DNABERT-2 fundamental model improves TF-binding predictions and provides insight into non-coding variations linked to disease.
Introduction
The genetic material that powers cellular activity, DNA, is converted into RNA and proteins. Transcription factors (TFs) regulate the transcription process, which is essential for gene expression. The makeup and operation of complete TF-DNA interactomes, especially for large mammalian genomes, are still poorly understood. A major challenge in genomics is identifying DNA sequence variations that change TF-binding sites, which can result in various human illnesses. A significant number of these variations are located in non-coding areas of the human genome and are essential for controlling the expression of certain genes.
Chromatin immunoprecipitation sequencing (ChIP-seq) is a crucial technique for analyzing particular TF-DNA interactomes. The immunoprecipitated DNA fragments must be sequenced using a particular anti-TF protein antibody to do this. Nevertheless, ChIP-seq has drawbacks, including costly deep sequencing, the possibility of mistakes from chemical TF-DNA fixation, and challenges with large-scale studies because of tissue and cell availability constraints and antibodies to TF posttranslational modifications.
Study Overview
The goal of the project is to create a foundation model based on DNA sequences and increase prediction accuracy by utilizing DNA breathing dynamics in a cross-attention aggregation module. To confirm the model’s resilience and adaptability, it was evaluated on various in vitro and in vivo datasets. The model considerably improved prediction performance in over 660 binding events out of 690, and it increased AUROC and AUPR measures by at least 3% on average, according to in vivo datasets. To provide a more sophisticated knowledge of TF-binding locations, the model also enabled cross-attention weights to identify evolutionary enriched motif regions. Additional tests on the SELEX dataset demonstrated that the breathing dynamics were more effective than sequence-only data, with gains of at least 3% on the R2 scale.
Key Contributions of the Study
- Utilizing DNA breathing dynamics to direct a foundation model to use various feature modalities.
- Incorporating the cross-attention framework into multimodal environments.
- Cross-attention weights’ explainability for identifying regions with enriched motifs.
- Extensive analysis of TF-DNA binding data both in vitro and in vivo.
Model Architecture of EPBDxDNABERT2
TFDNA binding affinities at certain chromosomal locations are determined using the EPBDxDNABERT2 computational model, which combines DNABERT-2 with EPBD computed properties of DNA breathing. Starting at location y on chromosome x, the model comprises a meticulously extracted DNA sequence of length l. To create an extended complete context sequence with a total length of L = f + l + f, an adjacent flank of predetermined size f is incorporated at both ends of the primary sequence.
More about EPBDxDNABERT-2
The attention weights of the EPBDxDNABERT-2 model were thoroughly examined and contrasted with randomized attention weights. Singular values for genuine EPBD traits declined more sharply, according to the data, indicating a concentration on fewer predictive features. On the other hand, randomized features showed a more gradual fall in the distribution of singular values, indicating a more distributed focus on less informative traits. This increased emphasis on EPBD features is interpreted as the precision of the model, demonstrating its improved capacity to identify and highlight the most predictive features for the prediction of transcription factor-DNA binding throughout the entire genome.
Similar to DNABERT-2, the EPBDxDNABERT-2 model uses a mechanism to extract conserved patterns from DNA sequences. Cross-attention values are used in this model to evaluate the impact of pyDNAEPBD characteristics. By computing average cross-attention weights along tokenized l tokens and the breathing dynamics of center 200 nucleotide bases, the study is predicated on a cross-attention matrix. This enables a more comprehensive contextual understanding of regions that bind and those that do not. The TOMTOM motif comparison tool is used to confirm the regions rich in motifs, which are identified using the weights, against the JASPAR CORE (2022) non-redundant experimental motif database.
Various species, particularly mouse TFBS prediction tasks, were used to assess the EPBDxDNABERT-2 model.
Using the mouse dataset from the Genome
Understanding Evaluation (GUE) benchmarks, the model outperformed the competition on four of the five tasks, with the MelMafkDm2p5dStd TFBS prediction task showing the most significant improvement of 4%. This work shows that the suggested model is universal in predicting TFBS across the whole genome.
Conclusion
Research has demonstrated the usefulness of the EPBDxDNABERT2 model in predicting TF-DNA binding sites. The model enhanced the accuracy of binding site prediction by combining DNA breathing properties from the EPBD nonlinear DNA mechanical model with the sophisticated DNABERT-2 foundation model. This multi-modal strategy provides a fresh viewpoint on genomic interactions by combining biophysical and computational information. The study emphasizes the complementary contributions of various EPBD feature types and the significance of cross-attention in feature integration. A significant advancement in computational genomics is represented by the model’s adaptability to a variety of datasets, which allows for a more thorough examination of intricate biological processes.
Article Source: Reference Paper | Reference Article | For access to the source software, visit the following GitHub repository and https://zenodo.org/records/11130474.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.