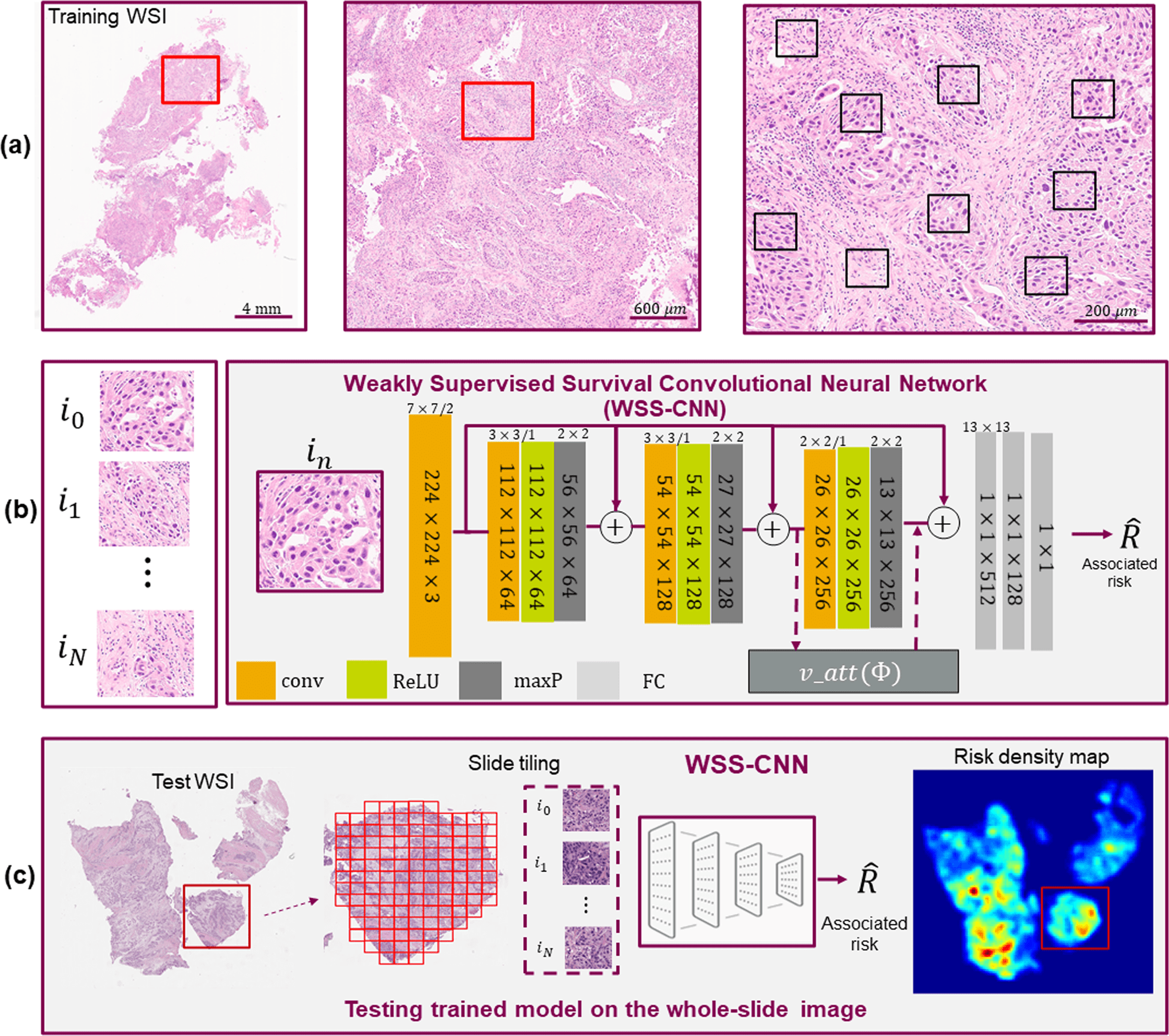
Scientists used hematoxylin and eosin (H&E) images with a deep learning approach to predict disease outcomes, an emerging technique for the improvement of clinical trials via computational methodology.
For treatment choices and disease outcome monitoring, it is highly clinically relevant to understand the variables that affect a cancer patient’s prognosis.
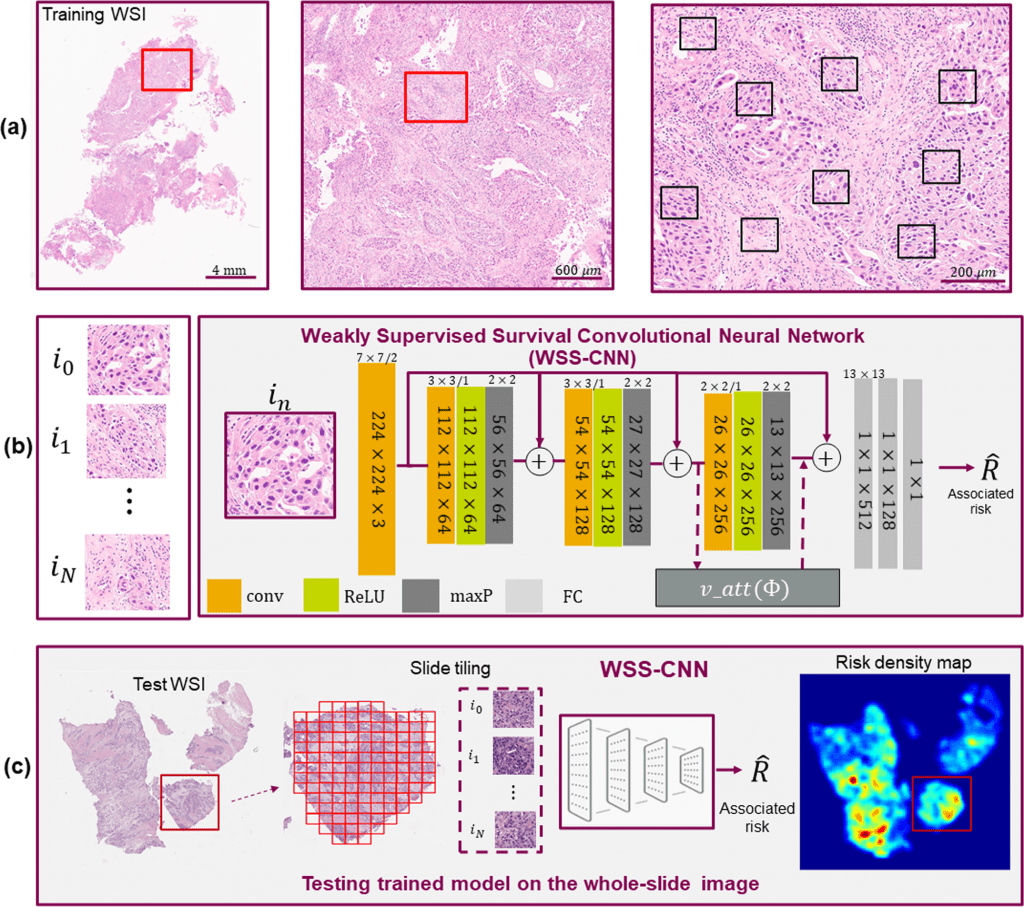
Image Source: Usability of deep learning and H&E images predict disease outcome-emerging tool to optimize clinical trials
The use of whole slide images (WSIs) of tumor tissue stained with hematoxylin and eosin (H&E) for accurate prognosis and prediction of response to targeted therapy presents an interesting possibility thanks to developments in artificial intelligence (AI) and digital pathology.
Effective training of AI models frequently necessitates hand-delineated annotations, which may not be easily accessible for more extensive data sets.
In this study, the scientists explored whether AI models could be trained only on patient-level survival data without using region-level annotations.
To predict overall survival, the researchers introduced a weakly supervised survival convolutional neural network (WSS-CNN) method incorporating a visual attention mechanism.
The addition of visual attention offers perceptions into areas of the tumor microenvironment with the pathological interpretation, perhaps enhancing their comprehension of the pathomechanism of the disease.
The scientist conducted this study on two separate, multi-center patient data sets for bladder urothelial carcinoma and lung cancer, both of which were publically available data. WSS-CNN features are indicative of overall survival in both tumor indications, according to our univariable and multivariable analyses, which they demonstrated.
The given results support the use of computational approaches to increase the effectiveness of clinical trial studies and emphasize the usefulness of computational pathology algorithms for prognosis prediction using just H&E stained images.
Influence of Components of TME Structure on Tumorigenicity
Clinical judgment and treatment planning for cancer patients are significantly influenced by survival (or time-to-event) analyses. Using biomarker/prognosis data, clinical trials in cancer divide patients into several treatment subgroups and assess survival in these groupings.
Monitoring survival rates or the amount of time it takes for a disease to advance can also reveal the effects of a patient’s reaction to a particular therapy or treatment.
Clinical professionals may benefit from objective survival probability estimation in personalizing treatment plans and improving participant recruitment for successful clinical studies.
Genomic and protein (immunohistochemistry, IHC) biomarkers are typically used in survival analysis, along with other clinical and patient characteristics or demographic data, such as age, gender, BMI, ethnicity, etc.
The tissue-based prognosis of the tumor microenvironment (TME), which is composed of composite structures of normal, malignant cells, connective tissue infiltrated with immune cells, and vasculature, is another recently developed but promising area.
There is mounting evidence that suggests that TME structural elements affect tumorigenicity and have been implicated in cancer prognostication.
Imaging-Based Prognosis is Attracting Attention
A skilled histopathologist manually examines cancer tissue slides under a microscope in standard procedure.
By examining the morphological differences inside the cancerous regions, measuring the density of malignant areas, and observing the spatial arrangement of the TME, the visual examination’s main goal is to identify cancer.
However, thorough visual inspection of tissue slides requires a lot of time and resources, and because histological methods are subjective, there will always be variation between and even within observers.
Artificial intelligence and machine learning (AI&ML) algorithms can now be used to predict pertinent clinical outcomes from histology image data, thanks to recent developments in computational pathology.
Automated algorithms are frequently built on digital image analysis principles, which can examine images to increase the accuracy and reproducibility of cancer diagnostics.
Artificial intelligence has the potential to identify intricate relationships between histology data and patient outcomes when combined with whole slide imaging (WSI) data sets comprising tissue pictures stained with hematoxylin and eosin (H&E).
The use of AI in immuno-oncology (IO) to measure tumor mutual burden (TMB) and PD-L immunohistochemistry (IHC) are related applications. Imaging-based prognosis is receiving increased attention as a result of the development of deep learning and the accessibility of scanned tissue slides.
Highlighting the Prognostic Significance of Components of TME
Several issues may hamper the development of these algorithms for standard clinical practice despite the enormous growth in deep learning-related technologies for histopathology image processing.
To train AI ML models and forecast the slide label, computational pathology algorithms often need correctly characterized tissue areas.
There are no specific annotations about which regions of interest (ROIs) from the tissue slides are more likely to impact survival in most real-world problems and for the task at hand. Instead, the ground-truth (GT) label for overall survival is typically provided at the patient level.
Selecting ROIs of a WSI (mostly from tumor component areas) recognized by a pathologist or via a third-party application and training a supervised learning model to predict survival are the most widely used methodologies currently in use.
One significant drawback of such systems is that they only learn the prognostic features from annotated ROIs, which always introduces bias into the model predictions.
Consequently, nothing is known about the prognostic importance of the complete TME spatial architecture. Another potential drawback is reusability, as it would not be possible to obtain finely detailed annotated portions of gigapixel images for training the model for larger data sets.
This supports the scientists’ prediction that to emphasize the predictive relevance of various TME components using H&E stained WSIs, AI models can be trained purely on patient-level survival data and without regions-level annotations.
A Deep Learning Framework for Leveraging Patient-Level Survival Statistics
In this study, the scientists provide a deep learning framework for predicting overall survival (OS) without the necessity for properly annotated regions by using patient-level survival statistics.
Without prior knowledge of tissue composite structures, their system illustrates the applicability of poorly supervised learning and visual attention mechanisms to infer pertinent spatial signals for predicting cancer outcomes.
This strategy might enhance their comprehension of the disease’s pathogenetic mechanisms and result in an unbiased, objective prognosis.
The researchers show the value of their prediction approach on two cohorts of multi-center data sets, including data on urothelial bladder carcinoma and lung cancer from the publicly available National Lung Screening Trial (NLST) data collection.
A preliminary assessment of the applicability of the proposed framework to various tumor indications is provided by the varied TME characteristics of the respiratory (lung) and urinary (bladder) systems.
By cross-validation of both data sets, they separately reported the prognostic accuracy of the framework. Overall, the proposed framework outperforms previously published approaches to reach state-of-the-art performance in OS prognosis on the NLST data and produces statistically significant results for the bladder cohort.
The researchers furthered their investigation through univariable and multivariate analysis to assess the predictive importance of various clinical and pathological parameters.
Last but not least, to better understand the course of the disease, the researchers obtained WSI density maps to explore regions of TME that may have predictive traits and connect with pathological interpretation.
Explainability is the ability to present findings so that human experts can understand the reason behind a decision. Without any presumptions or annotations, the WSI density maps show key danger locations that are consistent with pathologist interpretation.
The Endpoint
The given work introduces a new deep learning technique that might help with clinical trial design optimization but also has certain drawbacks.
The model is tested on random samples taken from WSIs; automatic, semi-automated, or some curriculum-based learning might be investigated for the selection of ROIs to test the model (for example, how many tissue areas to select from each slide).
The performance of the model on larger cohorts, especially with a homogenous distribution among different stages of the disease, may be validated as part of further extension of the WSS-CNN architecture.
Before using the WSS-CNN framework as a standalone clinical tool, prospective validation of the framework in clinical trial studies would be essential.
One possible application is to use this in clinical trial settings to categorize individuals as fast (high risk) and slow progressors (low risk) based on their disease results.
As a result, clinicians would be better able to assess the treatment response, pharmacological efficacy, and safety across various sub-groups and choose pertinent people for multi-arm clinical trial research.
The scientists believe that the suggested framework has the potential to be used with other medical imaging modalities in addition to various tumor indications for disease prognosis using deep learning and images. This discovery will probably pave the way for the creation of other AI-based imaging techniques for clinical outcome prediction and clinical trial design optimization in precision medicine.
Article Source: Qaiser, T., Lee, CY., Vandenberghe, M. et al. Usability of deep learning and H&E images predict disease outcome-emerging tool to optimize clinical trials. npj Precis. Onc. 6, 37 (2022).
DOI: https://doi.org/10.1038/s41698-022-00275-7
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.