About 20% of human proteins lack recognized functionalities, and over 40% lack context-specific functionals, underscoring the difficulties of comprehending these proteins and their varied roles in different states, illnesses, and situations. To model, generate, and predict protein phenotypes across five interconnected knowledge categories—functional protein domains, illness connections, therapeutic processes, molecular functions, and molecular interactions—researchers from Harvard Medical School, MIT, and King’s College London offer PROCYON and PROCYON-INSTRUCT is a comprehensive database with 33 million instructions for protein phenotypes. Combining protein and phenotypic data generates free-form text phenotypes in a single, cohesive model and achieves zero-shot task transfer. PROCYON makes it possible for proteins associated with small molecules to be conditionally retrieved, producing candidate phenotypes for poorly understood proteins linked to Parkinson’s disease.
Introduction
One of the main problems in biology is deciphering molecular phenotypes, which necessitates knowledge of molecular mechanisms governing living systems. Forty percent of human proteins miss context-specific functional insights, and about twenty percent lack clearly characterized roles. As only 5,000 well-studied proteins are the subject of 95% of life science papers, this bias in protein research further widens the knowledge gap. Multiple-scale protein phenotypes include molecular processes, therapeutic interactions, cellular pathways, organismal characteristics, and illnesses. The methods used today to predict protein function frequently fail to account for the entire range of phenotypes or forecast functions that are too imprecise to yield useful phenotypic data. The goal of projects such as the Human Proteome Project Grand Challenge is to identify the phenotypes of each protein in human health and disease, both individually and within networks and pathways.
Understanding PROCYON
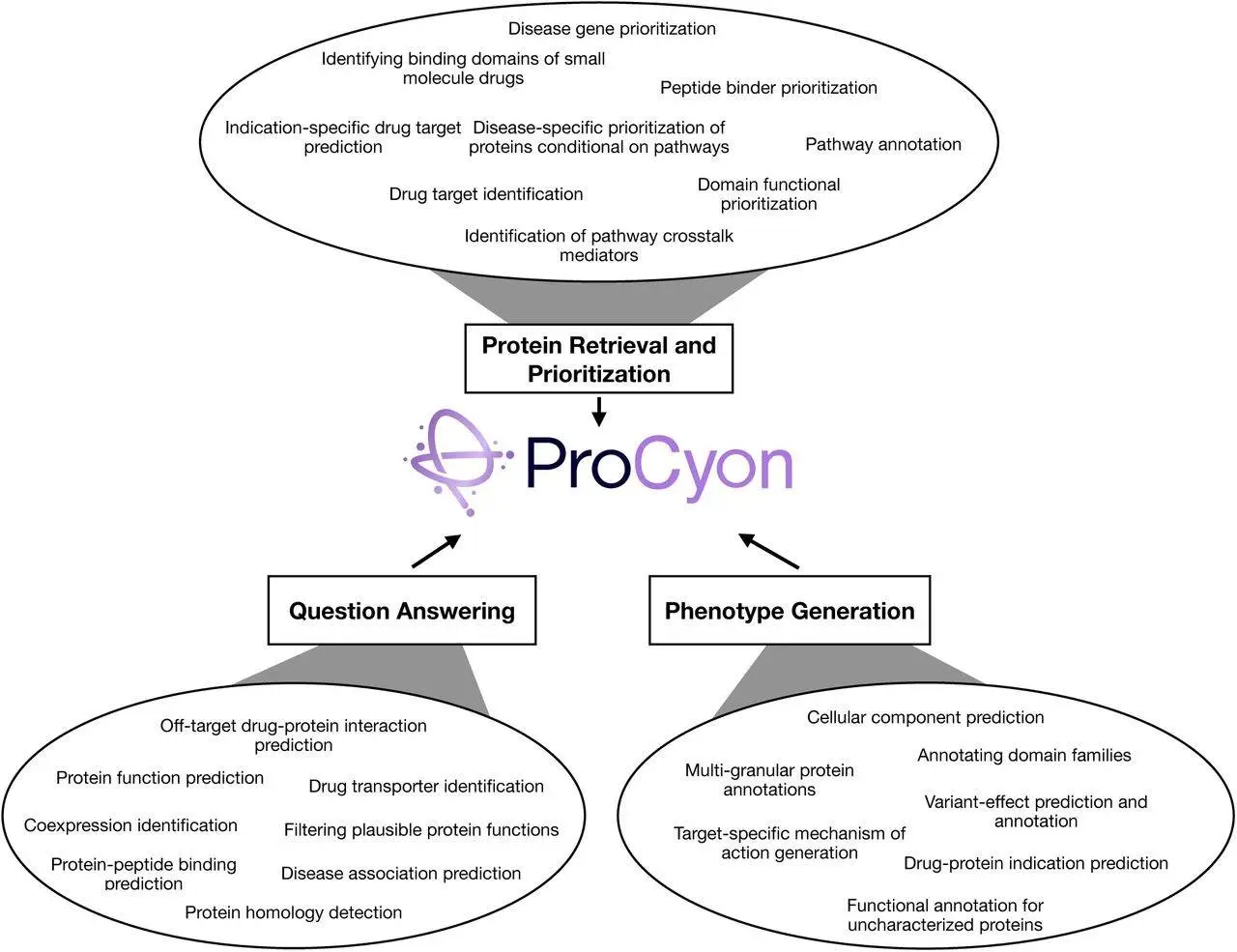
Protein phenotypes are modeled, generated, and predicted using the multimodal model PROCYON in various knowledge domains. The PROCYON-INSTRUCT instruction tuning dataset, which comprises more than 33 million human protein phenotypes, unifies these domains by integrating proteins with interleaved phenotype descriptions. To process arbitrarily interleaved inputs of text, protein sequences, domains, peptide data, and small molecule structures, the model co-trains a huge language model with specific encoders for protein sequences, structures, and medicines. Contextual protein retrieval and accurate phenotype predictions are made possible by PROCYON’s use of instruction tweaking on PROCYON-INSTRUCT to produce free-form text interspersed with recovered protein-related elements.
Application of PROCYON
The potent tool PROCYON predicts and generates phenotypes on various scales by modeling protein phenotypes in five interconnected domains. Protein-binding domains can be predicted with high precision by integrating sequence and structural data, identifying pathways and processes related to disease biology, and precisely generating molecular phenotype descriptions for poorly defined proteins.
It can retrieve proteins conditioned on small-molecule medicines, find peptides that bind target proteins, bridge molecular-scale phenotypes with more general disease-specific phenotypes, and offer mechanistic insights into understudied proteins thanks to its zero-shot task transfer capacity. One of PROCYON’s noteworthy advantages is its capacity to produce and predict phenotypes on various scales.
PROCYON enables researchers to study proteins in a contextualized manner by simulating and producing complicated protein behaviors. Its natural language interface makes these phenotypes accessible to a wider range of researchers, allowing for a more thorough investigation of proteins. The capacity of PROCYON to clarify illness contexts in input prompts, showcasing its ability to process composite questions, including medications and diseases, demonstrates its usefulness in drug-target discovery, including bupropion.
Limitations
PROCYON, a biological research tool, has to be improved due to its limitations. It may be unable to record complex biological circumstances for specific requests, even though it can produce written descriptions. It is difficult to evaluate novel phenotypes since there is a dearth of ground truth data. PROCYON’s inability to adapt to changing biological studies stems from its static character. Conversational features, which could allow for an iterative, fast improvement in multi-turn exchanges with scientists, are not yet included. Furthermore, PROCYON’s predictions do not quantify uncertainty, which may be crucial for researchers to consider when analyzing forecasts. Although PROCYON is not yet integrated with multi-omics datasets, this could improve its capacity to associate cellular settings with protein phenotypes in the future.
Conclusion
The multimodal character of protein representation and the complexity of protein phenotypes are two essential facets of human protein biology that PROCYON, a multimodal foundation model, aims to address. PROCYON facilitates insights that surpass pre-established phenotypic categories by bridging the gap between molecular data and human-readable phenotype descriptions by integrating protein phenotypes across many scales and knowledge areas. Because of this capability, PROCYON is positioned as a fundamental tool for enhancing the comprehension of protein phenotypes. Molecular functions, therapeutic mechanisms, disease connections, functional protein domains, and molecular interactions are the five knowledge fields that PROCYON, a multimodal model, focuses on simulating complex protein phenotypes. It allows for the creation of new insights by bridging the gap between molecular modalities and phenotypic descriptions. PROCYON connects phenotypic descriptions with molecular modalities by evaluating millions of human protein phenotypes. This allows for zero-shot task transfer, enabling the model to generalize to novel phenotype tasks not encountered during training. PROCYON can investigate phenotypes in an open-ended, hypothesis-driven fashion thanks to this methodology.
Article Source: Reference Paper | ProCyon’s model weights and codebase are available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.