A recent study by a team of researchers has made significant strides in unraveling this complex puzzle of human interaction. The human interactome encompasses all the protein-protein interactions (PPIs) that are essential for protein function and remains an outstanding challenge. It is critical to understand these interactions to understand disease pathology and new effective interventions. Studies seeking PPIs have relied heavily on traditional experimental methods, including yeast two-hybrid assays and affinity purification mass spectrometry. However, these methods have shortcomings, above all, high false positive and false negative rates, which limit the capacity of generating a comprehensive interactome.
Computational Approaches to the Rescue
With the advancement of science, computational approaches have become alternative ways of solving the problems posed by experimental studies. Since PPIs are evolutionary-based interactions, computational approaches help study PPIs by analyzing the sequences and structures of proteins to forecast potential based on evolutionary relationships, structural compatibility, and functional associations.
With the integration of computational tools in biology, there has been a great development in PPI prediction. Deep learning algorithms, in particular, have proven to be highly effective in identifying coevolutionary signals between proteins, which indicate potential interactions. On top of all, the volume of large-scale protein structure databases has provided valuable information for predicting PPI interfaces.
Key Challenges and Opportunities
Although these advancements are impressive, there are difficulties in predicting PPIs for several reasons:
The complexity of the Human Proteome: The human proteome contains 20,000 proteins and can be exceptionally complicated. This makes it hard to deduce all of the putative interactions between the proteins.
Dynamic Nature of PPIs: PPIs are not static; they can change in response to cellular signals, environmental conditions, and disease states.
Experimental Restriction: The experimental approaches for PPI identification are usually very tedious and costly, hence limiting the mass of data at hand.
However, these challenges also bring opportunities for new tool creation. By developing more accurate and efficient computational methods, researchers can address the drawbacks of experimental methods and make a whole picture of the human interactome.
Enhancing PPI Prediction: The Power of Deeper MSAs and Novel DL Networks
The researcher’s work aimed to improve the accuracy of PPI prediction by employing two main approaches:
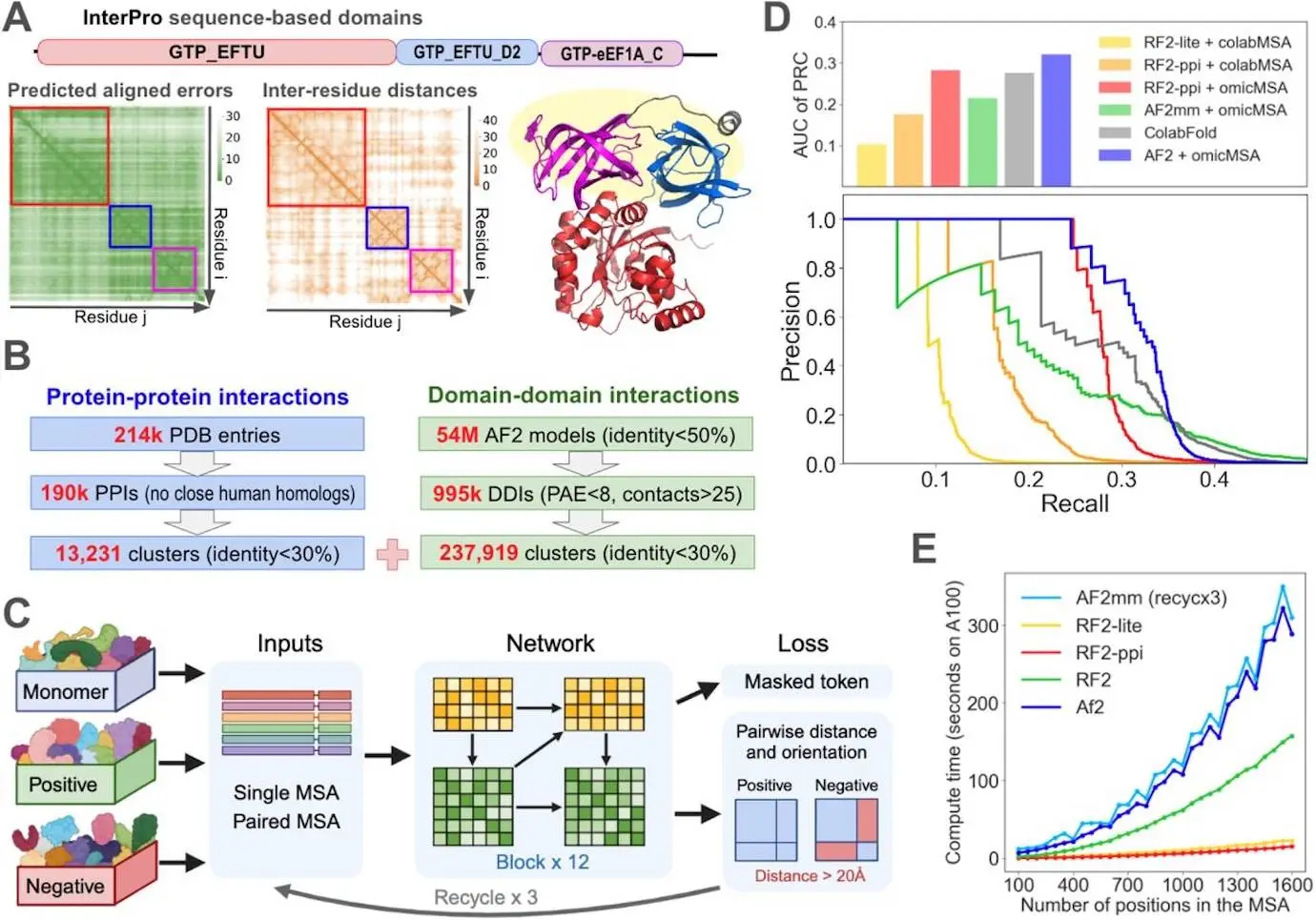
Enhancing Coevolutionary Signals with Deeper MSAs: The researchers obtained deeper multiple sequence alignment (MSA) from large MSA datasets obtained from 30 petabytes of unassembled genomic data. These deeper MSAs also include sequences from a wider range of species having more evolutionary signals, and more subtle evolutionary relationships are captured.
Developing a New DL Network Trained on Augmented Datasets of Domain-Domain Interactions: Furthermore, a novel deep learning (DL) network capable of learning on a new domain-domain interaction (DDI) was developed. DDI is the interaction between protein domains, which are distinct structural and functional units within proteins. Because DDIs can infer PPIs, training the DL network on such DDIs enhanced its ability to predict PPIs.
Key Findings from the study:
A deeper explanation of the approach adopted by the researchers gives rise to the following key findings:
Enhanced Coevolutionary Signatures: The coevolutionary hypothesis was useful in enriching deeper sequence data derived from multiple sequence alignments, enhancing interaction through the prediction of protein-protein interactions (PPIs).
Novel Deep Learning Network: The novel deep learning network RF2-ppi specifically includes features designed for interaction prediction and integrates MSA, inter-residue interaction, and 3D structural information.
Comprehensive PPI Identification: The current systematic investigation screened 200 million human protein pairs and obtained 18,316 PPIs that would be expected to be populated with 90% precision.
Novel Interactions: Of the above predicted protein-protein interaction, 5578 were novel predictions, expanding our understanding of the human interactome.
Biological Insights: The predicted PPIs provided valuable insights into protein function, cellular processes, and disease mechanisms.
Conclusion
The researcher’s work represents an important advancement in analyzing the human interactome. However, still there is a lot to be done. In the future, this predicted extension may include improvements in the prediction accuracy and extension of the PPI prediction scope to other organisms, a combination of experimental and computational data, and studying the effects of PPIs. As more data about the interactome are accrued, one can anticipate new cutting-edge discoveries and new therapeutic applications of the interactome. Predicting the interactions in molecular networks will help build the understanding of disease pathogenesis at the molecular level and assist in the design of appropriate therapeutics.
Article Source: Reference Paper | Predictions and intermediate results are available at http://prodata.swmed.edu/humanPPI.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.