Enzymes that have been specially engineered can improve the application of biocatalysts in industrial biotransformations and help address the biotechnological problems of the twenty-first century. The rotamer inverted fragment finder–diffusion (Riff-Diff) approach, which scaffolds catalytic arrays in de novo protein backbones with customized substrate pockets, is a hybrid machine learning and atomistic modeling technique presented by researchers. Researchers utilized Riff-Diff to build a catalytic tetrad that can effectively catalyze the retro-aldol process. High fold variety is shown in functional designs, which have substrate pockets resembling those of natural enzymes. A few of the resulting de novo enzymes exhibit activity comparable to those that have been refined by in vitro evolution. The design approach can theoretically be used with any combination of catalytically capable amino acids. These discoveries clarify basic concepts of enzyme catalysis and open the door to addressing issues related to the de novo protein catalysts’ practicability in industrial processes.
Introduction
Chemists use high-throughput screening as a vital method to find naturally occurring enzymes that exhibit the appropriate activity during chemical transformations. However, locating these enzymes calls for a lot of resources and screening power. Biocatalysts for diverse purposes have been developed through protein design; nevertheless, the significance of low initial catalytic rates has been overlooked. Directed evolution and high-throughput screening make up for this in the current paradigm. This method, however, is not appropriate for developing new, effective catalysts for chemical reactions that cannot be accessed by high-throughput screening. By addressing methodological flaws in functional design and enzymatic activity prediction, researchers can improve efficiency and expand the knowledge of the fundamentals of enzyme catalysis.
What is the De Novo Enzyme Design Model?
According to the transition-state model of enzyme catalysis, chemical processes can be accelerated by functional groups present in the active site, which stabilize the transition state of the reaction relative to the ground state. By arranging amino acid functional groups in a stabilizing shape around a computational model of a transition state, this concept enables the creation of minimum active sites or theozymes. The first computationally created enzymes were inserted into natural protein cavities to support the desired geometry. Although these enzymes had poor catalytic rate accelerations and frequently low success rates, they were able to successfully catalyze chemical reactions. The main limiting factor was the accuracy with which catalytic side chains could be placed in the active site, with successful designs exhibiting less than 1 Å side chain RMSD between the geometry in the theory and a high-resolution experimental structure.
Understanding Riff-Diff
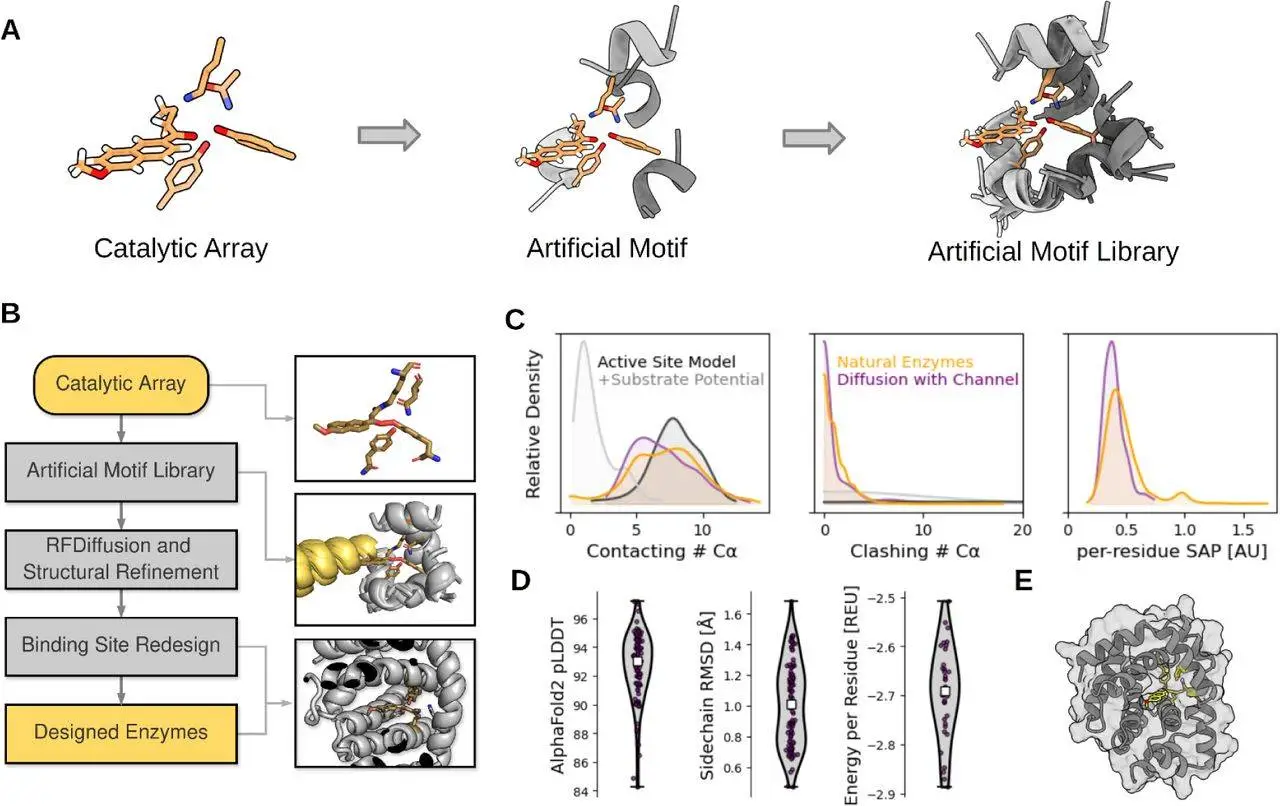
Researchers have developed a method called Riff-Diff (rotamer inverted fragment finder – diffusion), which focuses on creating efficient biocatalysts by incorporating an optimized catalytic center in de novo protein backbones. This method involves the precise placement of idealized backbone-side chain fragments, preorganization of given catalytic arrays, and the design of backbones with custom binding pockets. Researchers tested the effectiveness of this approach by designing retro-aldol enzymes based on a highly optimized catalytic tetrad. Detailed biochemical, biophysical, and structural characterizations and molecular dynamics simulations were used to test hypotheses about active site atom placement, preorganization, backbone-side chain compatibility, and enzyme conformational dynamics.
Riff-Diff in Artificial Motif Libraries Scaffold Catalytic Arrays
The structural preorganization of catalytic arrays is critical for promoting activity and achieving large catalytic rate accelerations. Rotamers that are suitable with the phi-psi angle combination of the fragment backbone are chosen in order to embed certain amino acids into idealized alpha-helical fragments. By doing this, the energy penalty connected with the favored catalytic rotamer is reduced, and the optimal backbone-side chain combination is guaranteed. Next, the fragments are assembled into fake motifs, which can be scaffolded with motif-scaffolding programs such as Chroma or RFdiffusion.
When using artificial motifs in motif-scaffolding with RFdiffusion, the size of the motifs that are chosen affects how successful the motifs are. Low scaffolding success rates are the result of these motifs being joined in arbitrary shapes determined by the placements of the catalytic array’s backbone atoms. A technique for creating and assembling libraries of synthetic motifs from catalytic arrays is devised to improve the success of scaffolding. To do this, rotamers of the catalytic array must be inverted, rotamers must be chosen for backbone compatibility with idealized helical segments, and combinations must be looked for that do not sterically clash. The rotamer probabilities of each active site residue are used to rank the non-clashing assemblies in an artificial motif library, which is then used to collect and store them.
Conclusion
Enzyme activity predictions have been essential to biocatalysis efforts aimed at producing superior catalysts with custom features for the production of sophisticated fine chemicals and novel medications. However, very few of these investigations were beginning from scratch, and those that were all depended upon directed evolution to improve the activities inside the biocatalysts that were constructed. Here, scientists developed a technique that has the potential to revolutionize enzyme design from a discipline requiring in-depth domain knowledge to one that the entire biotechnological community may utilize.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.