All important biological processes revolve around protein-protein interactions (PPIs’). Designing PPIs’ is difficult, nonetheless, because of the intricate structural characteristics that define them. Researchers propose BindCraft, an automated, open-source workflow with 10–100% experimental success rates for designing new protein binders. BindCraft generates nanomolar binders without the need for high-throughput screening or experimental optimization by using the deep-learning weights of AlphaFold2. The binders are made to target a variety of substances, such as multi-domain nucleases, allergens, and cell-surface receptors. These binders’ therapeutic and functional potential is illustrated, highlighting their prospective applications in biotechnology, diagnostics, and pharmaceuticals. This is a major step toward a “one design-one binder” strategy.
Introduction
Many biological processes, such as molecular recognition, structural support, and catalysis, depend heavily on proteins. These proteins depend on protein-protein interactions, or PPIs, to execute intricate functions like signal transduction, antibody-mediated immunity, and cellular communication. From a medicinal and biotechnological standpoint, it is highly promising to design protein binders that selectively target and modulate PPIs. These binders can suppress pathogenic agents, modify protein interaction networks, develop therapeutic antibodies, and produce biotechnological instruments for use in industry and research. Computational protein design provides a potent substitute for the time-consuming and tedious traditional ways of producing protein binders. Protein binders created from scratch have been employed to stop the spread of viruses, regulate the immune system and inflammation, stop the formation of amyloid, and regulate the processes involved in cell differentiation.
About Physics-based methods
Using scaffolding and sidechain optimization, physics-based techniques like Rosetta have played a significant role in early binder designs. On the other hand, these techniques have extremely low experimental success rates (usually less than 0.1%) and necessitate the creation and sampling of millions or even hundreds of thousands of designs. Furthermore, targets for such approaches may not be able to be targeted due to incompatibilities between the target and binder surfaces, which can lead to less-than-ideal binding interactions or even prevent the docking of preset scaffolds onto a fixed target structure.
Deep Learning and Protein Modelling
Recent advances in deep learning have greatly enhanced the ability to predict protein structure and the design of binders. Complex PPIs can be accurately modeled, and protein structures can be accurately predicted by models such as RoseTTAFold2 (RF2) and AlphaFold2 (AF2). By assessing projected complicated plausibility, AF2 filtering has led to a considerable rise in binder design success rates. Using RF diffusion for believable backbone generation and ProteinMPNN sequence generation, deep learning has also been successfully employed for de novo protein and binder design. On the other hand, this calls for a large amount of experimental in vitro screening in addition to thousands to tens of thousands of in silico designs.
Looking into BindCraft
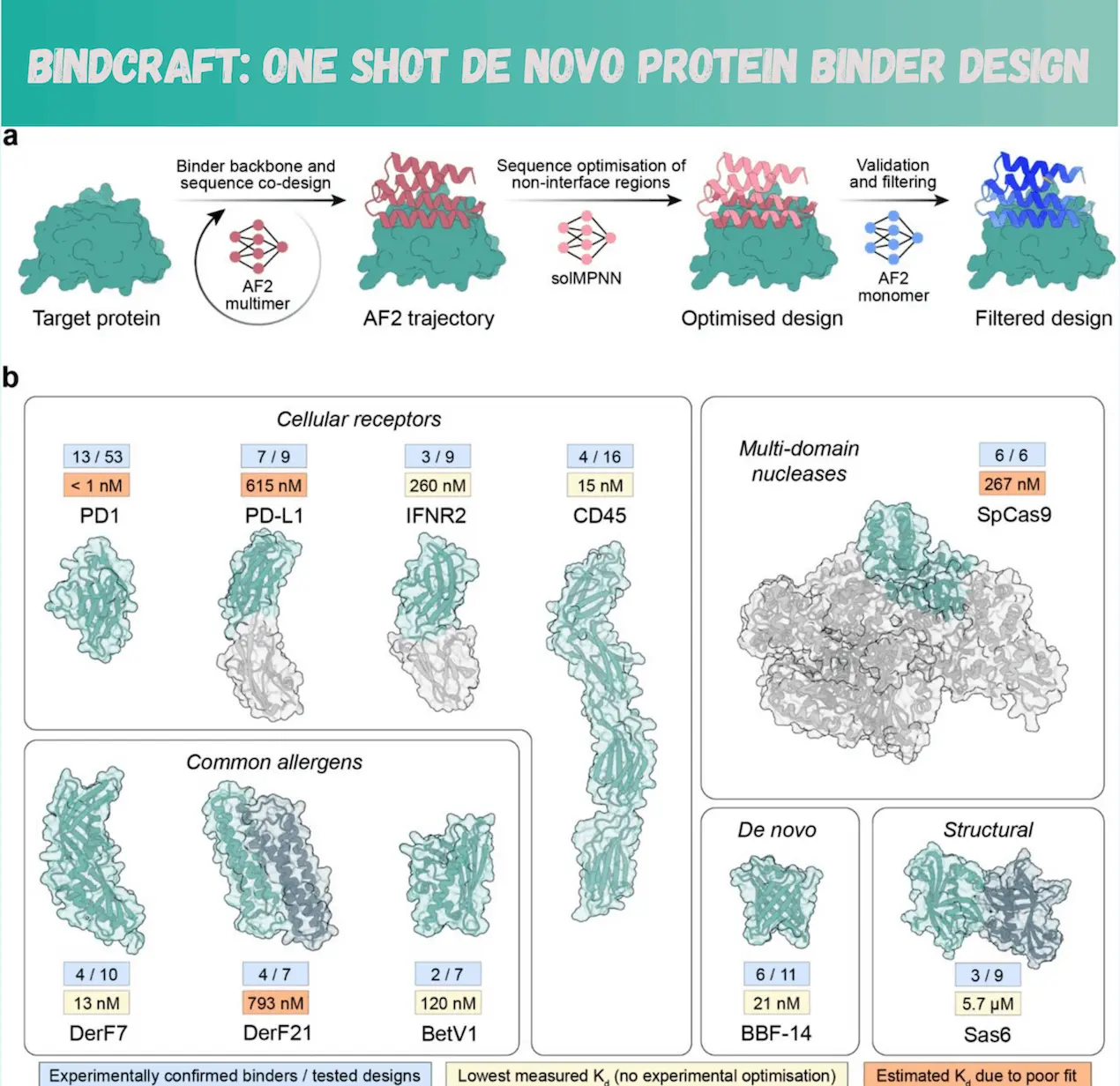
Using AF2’s training weights and protein structural patterns, BindCraft is an easy-to-use pipeline that creates protein binders. This revolutionary method effectively hallucinates novel binders and interfaces without requiring a large amount of sampling by using backpropagation over the AF2 network. Without requiring high throughput screening, the pipeline has been proven on ten different protein targets and has found multiple high-affinity binders. This is a significant development in protein design that opens up the field to research organizations without huge computer resources or high throughput screening capabilities.
Diving Deep into the Model
BindCraft is an automated pipeline that reduces the requirement for large-scale sampling by accurately designing binders using AF2 models. The user enters the desired designs, the size range of binders, and the target protein structure. The most efficient binding site is automatically chosen by the AF2 network based on the loss function. An error gradient is computed by the ColabDesign version of AF2, which backpropagates binder sequences using training weights. In order to fit certain loss functions and design requirements, the binder sequence is updated and optimized using this error gradient.
High-quality designs are produced through network iterations, allowing for the simultaneous creation of the binder structure, sequence, and interface. Every time, BindCraft co-folds the target and binder, giving both the binder and the target defined degrees of flexibility at the sidechain and backbone levels. This allows for creating interfaces and backbones that are shaped to fit the target binding site.
A model trained on protein complexes, AF2 multimer, is used to accurately model PPIs in constructing the first binders. All five trained model weights are used to prevent overfitting sequences. While keeping the interface intact, MPNNsol optimizes the binder core and surface sequences. Utilizing the AF2 monomer model, which has only been trained on monomeric proteins, the optimal binder sequences are predicted in order to reduce prediction bias and select high-quality interfaces. AF2 confidence metrics and Rosetta physics-based score metrics are used to filter predicted designs because deep learning models tend to provide unlikely outcomes.
Conclusion
BindCraft uses deep learning to comprehend chemical recognition determinants and improve protein design. This method makes the target protein’s side flexible, which is essential for efficient molecular recognition. Diversifying the structure in the direction of more intricate and natural folds is the goal to potentially produce binders based on nanobody technology and other pertinent molecular formats for clinical translation. The goal of the pipeline is to quickly generate binders for research, biotechnology, and medicines by getting to the point of “one design, one binder.”
Article Source: Reference Paper | Code is available on GitHub | Notebook for running BindCraft is available at Google Colab.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.