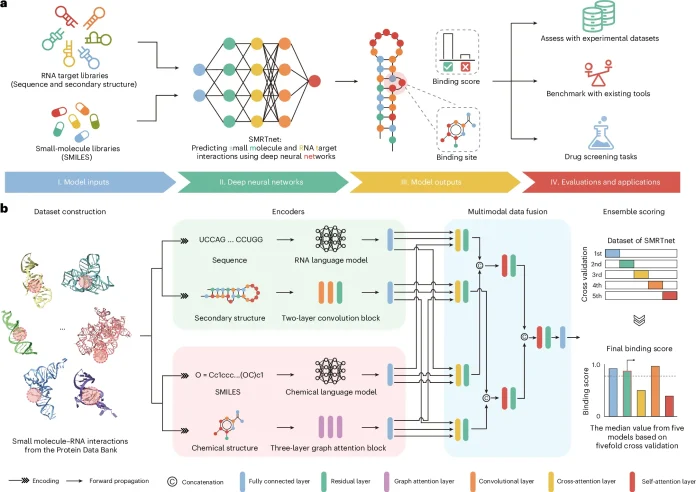
Researchers from Tsinghua University and Peking University in China developed the SMRTnet tool, which employs a deep-learning approach to identify interactions between small molecules and RNA, eliminating the need for RNA tertiary structure information. By combining RNA secondary structure and multimodal neural networks, the research showed that SMRTnet outperformed other prediction tools and validated its accuracy by identifying and experimentally verifying 40 small molecules that target disease-associated RNAs. The researchers tested their results by analyzing the MYC internal ribosome entry site (MYC IRES) and found that their predicted data could be confirmed via laboratory experiments, and one such compound reduced MYC expression, suppressed the growth of cancerous cells, and increased apoptosis.
Why is Finding RNA-binding Drugs Still a Challenge?
Drugs that bind to RNA and alter its function, such as splicing, translation, RNA-protein interactions, and viral replication, are known as RNA-targeting therapeutics. As observed by Evrysdi for spinal muscular atrophy, this enables the treatment of illnesses associated with undruggable proteins. Although wet-lab screening techniques like the Automated Ligand Identification System (ALIS), small-molecule microarrays, and biophysical binding assays experimentally test RNA-small-molecule interactions, they are slow and limited. While deep-learning models like RNAmigos2, RLaffinity, dry-lab tools like AutoDock Vina, RLDOCK, NLDock, and rDock carry out docking. Yet, the majority of methods depend on accurate RNA tertiary structures. Finding RNA-targeting small molecules is a major challenge for both experimental and computational discovery approaches because RNA takes on flexible, dynamic 3D shapes that are hard to determine and rare to come by.
SMRTnet: A Deep Learning Model
SMRTnet is a deep learning tool that can predict which small molecules can bind to RNA, even when the three-dimensional structure of RNA is unknown. Two large language models are used to learn patterns from RNA and chemical sequences. Convolutional neural networks (CNNs) help to identify significant local features, and graph attention networks (GATs) help to comprehend relationships within molecules. Multimodal data fusion (MDF) combines all of these features to produce precise predictions.
SMRTnet: From Predictions to Proof
Real RNA–small molecule binding structures from the Protein Data Bank were used to train SMRTnet. After cleaning the data, researchers extracted 31-base RNA fragments around binding sites, paired them with their actual ligands, and produced negative pairs. Because the dataset was divided, no molecule from the test set appeared in training. SMRTnet outperformed RNAmigos2 with a strong performance.
Even after eliminating similar molecules or RNAs, performance remained high, indicating that the model was not memorizing the data. Accuracy decreased when ligand RNA pairs were purposefully mismatched, demonstrating that SMRTnet actually learns actual RNA-small-molecule interaction patterns. SMRTnet predicts which small molecules can bind to RNAs linked to disease. After scoring 7,350 natural compounds, the researchers selected a smaller, mixed group of 376 compounds (strong, medium, weak binders) for laboratory testing. In this manner, they could fairly assess the accuracy of SMRTnet’s predictions.
Using Grad-CAM and SmoothGrad to determine which regions of the RNA the model deemed significant and comparing these with actual experimental binding sites, the researchers first investigated whether SMRTnet could determine which nucleotides in an RNA actually bind small molecules. With auROC scores, SMRTnet matched the standard scores well, particularly for RNAs such as MYC IRES, HIV TAR, CAG repeats of HTT (Huntington disease), and pre-miR-155 involved in cancer.
The ten disease-related RNAs-MYC IRES, pre-miR-155, HOTAIR helix 7, HIV-1 RRE IIB element, the CAG repeat in Huntington’s disease, and five structured RNA elements from the SARS-CoV-2 5′UTR, were then screened against 7,350 natural compounds using SMRTnet. Using microscale thermophoresis, the top 20 compounds per RNA (a total of 190) were tested in the lab, and 40 of them demonstrated actual binding (~21% success). Six exhibited extremely strong nanomolar binding, while the majority bound with micromolar strength. This demonstrates that SMRTnet can locate actual RNA-binding molecules and identify their binding sites.
Using SMRTnet in Cancer Biology
To determine whether SMRTnet’s binding predictions align with actual biology, it was tested on the MYC IRES RNA, a cancer-related RNA that aids in the production of the MYC protein. Researchers used microscale thermophoresis (MST) to test 376 small molecules with varying SMRTnet scores. Of these, 15 actually bound MYC IRES; higher SMRTnet scores indicated a higher likelihood of true binding. SMRTnet predicted that irinotecan hydrochloride trihydrate (IHT), a compound with good drug-like properties, would bind a particular internal loop (5′-UUCG / 3′-ACCC).
To demonstrate this, they created 20 MYC IRES mutants that altered or eliminated this loop. When the loop was broken, both SMRTnet scores and actual binding decreased, confirming the binding site. IHT outperformed MYC-RiboTAC in cancer cells, reducing MYC mRNA by approximately 57%, MYC protein by approximately 72%, slowing cell growth by approximately 20–48%, and increasing cell death by approximately 57–124%. Additionally, a luciferase reporter demonstrated that IHT selectively inhibits MYC IRES activity. All things considered, SMRTnet not only predicts actual RNA-binding molecules but also identifies their binding sites and aids in the discovery of drug-like substances that inhibit RNAs that cause cancer.
Conclusion
A significant barrier in RNA-targeted drug discovery is addressed by SMRTnet, a deep learning model that predicts small-molecule RNA interactions without requiring RNA tertiary structures. It outperforms current methods and successfully identifies biologically active compounds, including molecules that inhibit MYC expression in cancer cells. By combining RNA sequence, RNA secondary structure, and chemical features, it was also demonstrated that the accuracy of SMRTnet is highly dependent on secondary structure and RNA sequence. Performance decreases when structural information or essential elements like the multimodal fusion module and RNA language model are removed.
Article Source: Reference Paper | Availability: Google Colab | Installed through PyPI or https://pypi.org/project/smrtnet-latest/.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Jainab Shaikh is a postgraduate in Biotechnology with a strong interest in understanding how research translates into real-world innovation. Her areas of focus include biosensors, bioinformatics, and sustainable biotechnological applications. She is passionate about exploring recent scientific advancements and communicating them through clear, engaging, and accessible content. Her work particularly emphasizes research-driven narratives in healthcare, biotechnology, skincare science, and emerging life science innovations.