AI is being used to find novel antibiotics for bacteria resistant to pandemics; however, the currently used techniques have serious drawbacks. While generative models design molecules—which are difficult to synthesize—property prediction models, which assess molecules individually, have difficulty scaling to enormous chemical spaces. The generative model SyntheMol, which creates readily synthesizable chemicals from a chemical space of 30 billion molecules, is presented here by researchers from Stanford University and McMaster University. Utilizing SyntheMol, compounds that inhibit the bacterial pathogen Acinetobacter baumannii were created. Six structurally unique compounds with strong action against Acinetobacter baumannii were among the 58 produced molecules that were synthesized and experimentally confirmed.
Introduction
In modern medicine, the global spread of antibiotic resistance determinants poses a serious concern, with a projected 4.95 million deaths attributed to drug-resistant illnesses in 2019. Acinetobacter baumannii, a Gram-negative bacterium, is considered a critical priority by the World Health Organisation. Promising medication candidates, including antibiotics, can be quickly and precisely identified using artificial intelligence (AI) techniques. Although property prediction AI algorithms are time-consuming for vast chemical spaces, they are capable of evaluating chemical libraries to determine compounds with desirable properties. In contrast, generative models construct molecules from the ground up by fusing together smaller molecules with desired characteristics. This approach allows for the direct design of potential molecules without the need for a lengthy review process involving numerous compounds.
The inability of generative models to synthesize chemicals and molecules is a significant drawback. The intractable nature of these produced chemicals makes experimental validation impossible. Modern methods are needed since, although in silico results have been encouraging, not much research has synthesized and evaluated produced compounds, particularly in the development of antibiotics.
Understanding SyntheMol
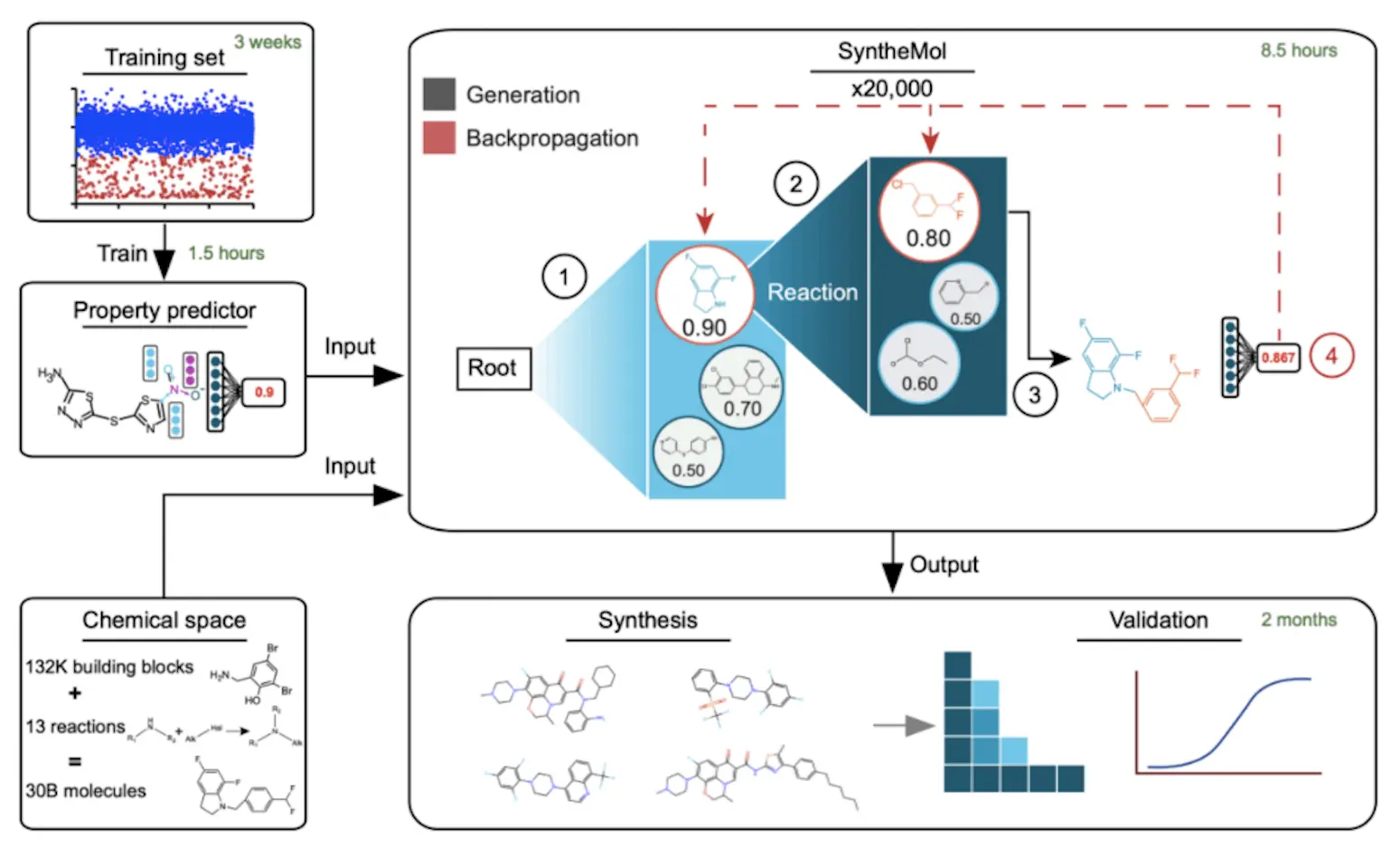
In this work, the researchers created SyntheMol, a generative artificial intelligence model that assembles unique compounds utilizing around 132,000 molecular building blocks with known reactivities and 13 thoroughly verified chemical synthesis reactions through the use of a Monte Carlo tree search. Nearly 30 billion easily synthesized compounds can be found in the resulting chemical space, with synthesis success rates exceeding 80% in just three to four weeks. Six of the 58 created molecules demonstrated strong activity against A. baumannii, and numerous other phylogenetically different bacterial pathogens taught SyntheMol to design molecules with antibiotic activity against A. baumannii, then synthesized and experimentally validated the 58 developed molecules.
About Property Prediction Models
Researchers physically screened 13,524 chemicals to create a training dataset. Researchers then assessed the growth inhibition of A. baumannii ATCC 17978 after treating each chemical, yielding 13,054 inactive and 470 active compounds. Researchers developed three models to predict antibacterial activity against A. baumannii using this dataset. Chemprop is a molecular property prediction system that uses a directed message passing neural network (MPNN). An alternative to Chemprop, called Chemprop-RDKit, concatenates the MPNN embedding with a set of 200 molecular features that RDKit computes before the feed-forward layers. A random forest classifier called Random Forest feeds 100 decision trees with 200 RDKit features. Using divisions of 80% training, 10% validation, and 10% test data, researchers used 10-fold cross-validation to train each model type on the training dataset. With ROC-AUCs in the range of 0.80–0.84 and PRC-AUCs in the range of 0.35–0.40, all three model types performed equally after being trained on 16 CPU cores in less than 90 minutes. The researchers treated the models from the ten cross-validation folds as an ensemble of ten models throughout the creation of molecules.
Algorithm Used in SyntheMol
SyntheMol searches a large combinatorial chemical space for molecules with interesting molecular characteristics using a Monte Carlo tree search (MCTS). A property prediction model, which creates molecules using 132,479 building components and 13 chemical reactions, serves as the basis for the MCTS. A molecular property prediction model is then used to assess the molecule. Each phase of the search tree entails assigning a score to prospective nodes based on a system, which can represent individual building components or entire molecules. High-scoring molecules are given priority by the exploit score, whereas the node’s molecules’ property prediction score is influenced by the diversity penalty, explore score, property prediction score, and exploit score.
Filtering Antibodies with SyntheMol
The goal of the study was to identify structurally unique compounds for experimental validation that had good scores for property prediction. Three filters were created: 1,005 antibacterial chemicals from the ChEMBL database and Tversky similarity between Morgan fingerprints and active training set molecules. In order to guarantee structural uniqueness, molecules with a Tversky similarity > 0.5 were eliminated. The best twenty percent of prediction results were retained for effective compounds. Fifty clusters of molecules were subjected to Tanimoto distance-based k-means clustering in order to identify structurally different compounds. For experimental validation, the top-scoring molecule from each cluster was chosen.
Validation of Generated Molecules
In order to synthesize and validate the bioactivity of 150 produced compounds, the study used 26 molecules from Chemprop, 22 from Chemprop-RDKit, and 10 from random forest to synthesize 58 (83%) of the compounds in 4 weeks. Baumannii Growth inhibition tests against A. baumannii ATCC 17978, the strain utilized for training set curation, were used to validate these compounds. Six compounds with a minimum inhibitory concentration (MIC) of less than 8 µg/mL had significant antibacterial activity, as demonstrated by their 10% hit rate when coupled with either 1 MIC SPR 741 or 1 MIC colistin. Fifty-eight randomly chosen compounds from the Enamine REAL Space were examined as a control, but none of them showed any antibacterial action against A. baumannii.
Six compounds, including Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, A. baumannii, and Staphylococcus aureus USA 300, were tested by the ATCC against both Gram-positive and Gram-negative bacteria. Findings indicated that every chemical exhibited broad-spectrum antibacterial activity, with the possible exception of P. aeruginosa, whose cell membrane impermeability may be the cause of this.
Conclusion
A. baumannii and other ESKAPE species were targeted by antibacterial chemicals created using SyntheMol, a unique generative AI model. Using MCTS in conjunction with molecular property prediction models, 58 structurally unique and varied molecules were created and examined. By showing activity against A. baumannii and other phylogenetically diverse ESKAPE species, six of these compounds proved that generative AI might be used to effectively create small molecule antibiotic candidates.
Article source: Reference Paper | Reference Article | SyntheMol code is available on GitHub
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.