Sequence-to-structure predictions are the main focus of current computational methods for reverse translation, which is the process of translating complicated protein structures into sequences. This is how protein design is traditionally understood. The researchers from the InsAIght Research Center and the Public University of Navarra present StructureGPT, a cutting-edge deep learning model that translates complicated protein tertiary structures into relevant amino acid sequences using sophisticated natural language processing algorithms. Protein engineering is improved by StructureGPT, a revolutionary paradigm, especially in industrial and pharmaceutical applications. By producing amino acid sequences from intricate structural inputs, it enhances the design of proteins with particular functions. This model shows its usefulness in a variety of biomedical and biotechnological applications by predicting sequences with high accuracy and suggesting alterations for improved protein properties. It does this by making use of linguistic similarities between human language and protein structures.
Introduction
Proteins are vital to several biological processes owing to their diverse range of functions and present a plethora of opportunities for research, industry, and pharmaceuticals. Our capacity to create and modify these molecules for particular purposes has greatly increased with the development of computer models. With its remarkable ability to predict protein structures from amino acid sequences with previously unheard-of accuracy, AlphaFold2, a prominent example that was presented by Jumper et al. in 2020, has completely transformed the discipline. However, converting complicated protein tertiary structures back into sequences is still a challenging task that must be addressed, particularly in comprehending and modifying the activity of proteins.
Understanding StructureGPT
A deep learning model called StructureGPT interprets protein structures and produces related amino acid sequences using NLP techniques. This model can be used for various protein design tasks, which increases its usefulness in biomedical and biotechnological settings. With the help of StructureGPT, numerous amino acid sequences may be produced from the structure of a protein, and sequences can be optimized for particular functional characteristics like stability and solubility. In order to incorporate missing structural features during sequence prediction, autoregressive generation conditioned on specific structural information is employed. With this strategy, protein engineering has advanced significantly, and scientists can now produce unique protein variations with customized functions, opening up new avenues for sustainable environmental and medical advancements.
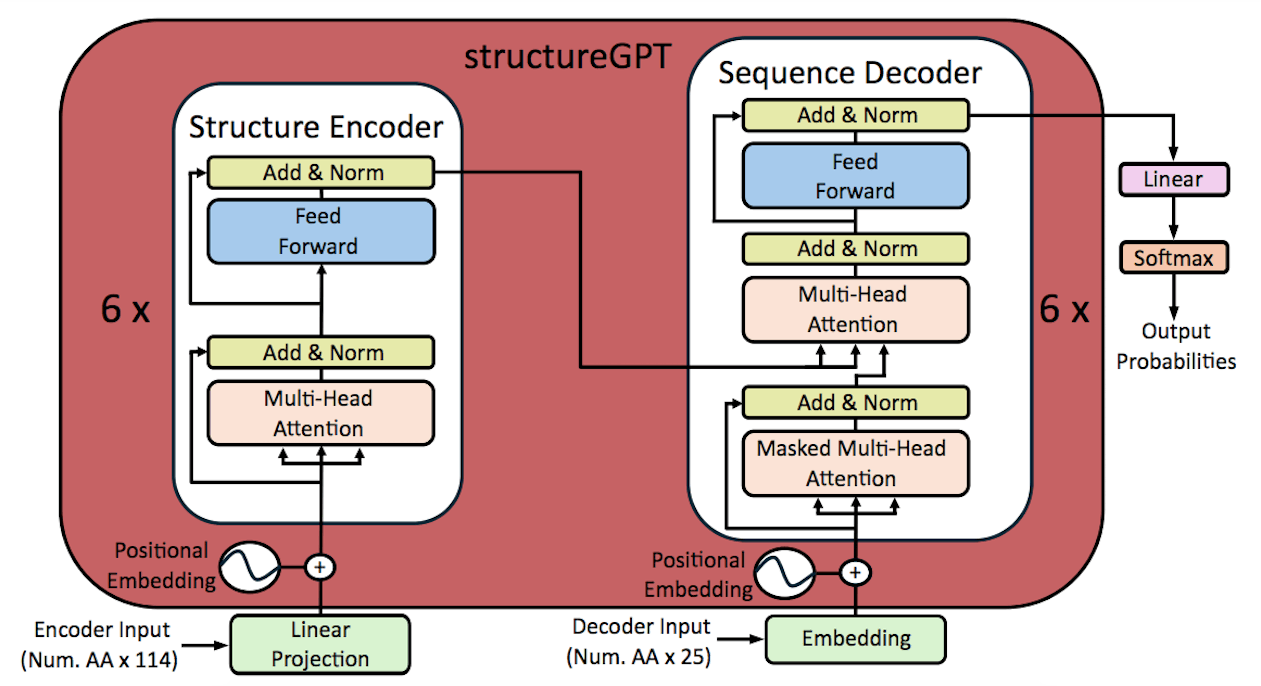
Model Architecture
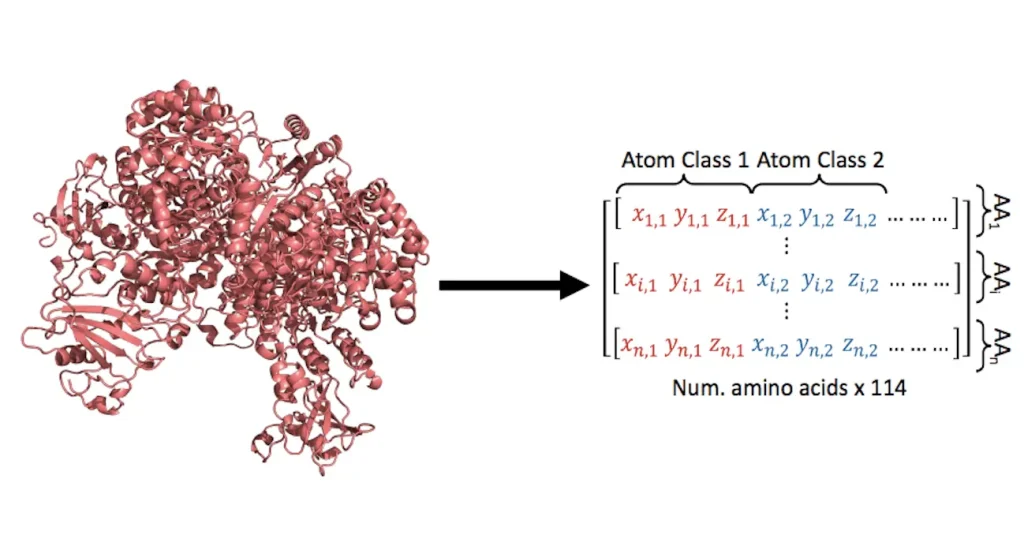
The translation problem in this work is quite different from traditional language problems that usually utilize indices and vectors inside a vocabulary. Retaining accurate structural information dictates the initial dimensionality of the vectors that represent amino acids. A linear layer was added in place of the word embedding block in the Transformer encoder-decoder architecture in order to attain larger dimensionality. A language model, which is usually necessary for natural languages, must assign a class to each word in order to solve the translation problem. To represent natural amino acids found in proteins in the structures-sequence translation problem, merely twenty distinct classes are required. This multiclass classification task was trained on StructureGPT, which was able to capture the sequence language in 20 different classes.
Training of the Model
The Transformer encoder-decoder was trained utilizing two datasets. Variations were introduced because of variances in the data used, even though both training processes were carried out in similar circumstances. CosineAnnealingLR was used to lower the learning rate, the AdamW optimizer was employed, and the Xavier uniform method was utilized to initialize parameters. A batch size of four samples was used for both trainings, which were run on two NVIDIA A100 computers with 40GB of VRAM. It took 37 epochs to finish the first training and 70 epochs to complete the second. In terms of data and learning rate, the training procedure was carried out autonomously.
Testing StructureGPT Generation
StructureGPT is an effective technique for producing amino acid sequences based on structural knowledge, which helps solve design challenges. With the help of its generative capabilities, which include the ‘greedy decode’ method, elements with the highest anticipated likelihood of appearing in a position can be added. Because of the model’s great accuracy during training, the sequences perfectly correspond with the original sequence. Nonetheless, it has been noted that the model has committed mistakes such as insertions and inversions. Errors, deletions, and inversions could be explained by the autoregressive scheme of the model. Insertions, deletions, and insertions are the most common faults found in StructureGPT. These errors might result in mismatches between the translated and original sequences. All things considered, StructureGPT’s generative features are quite helpful for completing design jobs.
Conclusion
A distinct atom encoding approach is used by the deep learning model StructureGPT to convert protein tertiary structures into amino acid sequences. This approach generates amino acid sequences autoregressively, which improves the design of proteins with specialized functions. The generative powers of the model are exemplified in its capacity to improve solubility and stability, two crucial characteristics in industrial and pharmaceutical settings. With the use of the model’s inpainting abilities, which demonstrate its comprehension of protein tertiary structure, a variety of protein sequences with structurally identical results can be produced. The model can’t, for example, predict functional results from sequences derived from structures or handle quaternary protein structures. Expanding the use of StructureGPT in functional genomics and proteomics should be the primary goal of future research.
Article Source: Reference Paper | StructureGPT’s source code is freely available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.