A team led by Karolinska Institutet used artificial intelligence (AI) and structural biology to learn more about two related proteins that protect the urinary tract and the gastrointestinal system from bacterial infection. The findings were published in “Nature Structural & Molecular Biology.”
“These findings could in principle be explored to develop a decoy alternative to conventional antibiotic treatments. And our structural work explains many human mutations associated with kidney diseases, as well as provides information on a protein region whose interaction with filtered light chains is implicated in cast nephropathy”, tells Professor Luca Jovine at the Department of Biosciences and Nutrition and corresponding author of the paper.
Image Source: https://news.ki.se/crystallography-cryo-em-and-alphafold-a-perfect-trio-sheds-light-on-key-human-antibacterial
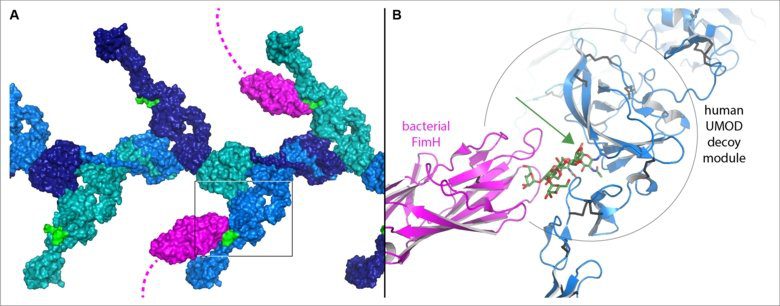
Uromodulin (UMOD) is a protein whose filaments help prevent urinary tract infection, the most prevalent non-epidemic bacterial infection that affects roughly 150 million individuals each year. It has a unique sugar chain that hasn’t been changed like others and is still high in mannose. This serves as a decoy by imitating high-mannose sugar chains on the urinary tract’s surface, which bacteria would ordinarily connect to with a sugar-binding protein called FimH at the tip of their hair-like appendages. Instead, the bacteria bind to UMOD’s sugar chain and are subsequently excreted in the urine. Evidence has emerged in recent years that glycoprotein 2 (GP2), a UMOD-like molecule produced in the pancreas and gut, serves a similar role in the gastrointestinal tract in combating bacterial infections.
“We wanted to determine which sugar chain of GP2 corresponded to the high-mannose chain of UMOD and understand how the proteins manage to keep these sugars ‘special’ so that they act as a bait for bacteria,” says Luca Jovine.
AI is establishing itself as a structural biology tool
The team collected X-ray crystallographic and cryo-electron microscopy (EM) data on GP2 and UMOD in collaboration with Daniele de Sanctis at the ESRF synchrotron in Grenoble and Marta Carroni at SciLifeLab in Stockholm. However, it was difficult to comprehend due to various technological problems. The scientists then began working with DeepMind and the team behind AlphaFold, a machine learning software that gained tremendous attention from the structural biology community thanks to its success in the 14th Critical Assessment of Structural Prediction (CASP) competition. Even though AlphaFold has never “seen” UMOD or GP2, its predictions were remarkably accurate. This enabled the team, which included Bin Wu of Nanyang Technological University in Singapore and Nao Yamakawa of Lille University, to decipher the GP2 X-ray data and fit low-resolution UMOD cryo-EM maps.
The research discovered that UMOD and GP2’s functionally critical decoy module comprises two pieces separated by a crevice. Although the sugar chains that bind bacteria are connected to distinct sections of the GP2 and UMOD sequences, the 3D structures demonstrate that their bases are positioned within the crevice in both situations. This explains how these specific sugar chains are protected from mutation and can thus be used as pathogenic bacteria fishing hooks.
Story Source: Stsiapanava, A., Xu, C., Nishio, S. et al. Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH. Nat Struct Mol Biol (2022). https://doi.org/10.1038/s41594-022-00729-3
Image Source: https://news.ki.se/crystallography-cryo-em-and-alphafold-a-perfect-trio-sheds-light-on-key-human-antibacterial
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar