The researchers at Broad Institute and Mass General Brigham in Nature Genetics propose DNA-Diffusion, the first ever generative diffusion framework trained on DNA accessibility data to design cis-regulatory elements (CREs) with activity specific to cell-types. The authors validated these synthetic DNA elements using STARR-seq and EXTRA-seq (Endogenous expression Targeted Regulatory Activity sequencing) against blood cancer B-cells, showing they can modulate endogenous genes.
Cell Type Specific Regulatory Elements: What Happens When Specificity Is Ignored
Cis-regulatory elements (CREs) in biological contexts are key short DNA sequences that control gene expression in cells, like enhancers, silencers, promoters, and insulators. They make up a large fraction of the human genome and act differently depending upon the cell’s identity, ensuring that genes are activated only in the right cells, at the right time, and in the right amount. For example, a liver enhancer activates genes for detoxification enzymes only in hepatocytes.
They work by binding transcription factors and interacting with chromatin structure. Making sure that each cell expresses only the gene it needs, or else normal physiology will be disrupted. An example can be enhancers meant for other genes, which activate MYC in T-cells, causing uncontrollable cell growth, leading to T-cell acute lymphoblastic leukemia.
Earlier approaches to regulatory elements in drug discovery and gene therapy relied on trial and error, motif libraries, or CRISPR-based enhancer editing, but these methods often showed low precision, scalability, and specificity based on cell type, which limits their therapeutic safety and effectiveness.
DNA-Diffusion, developed by an interdisciplinary collaboration, is promised to overcome these shortcomings by using generative AI for designing synthetic CREs with predictable, cell-specific activity.
As suggested by senior author Luca Pinello, synthetic CREs could be combined with existing gene therapies to make sure therapeutics reach the correct tissues, reducing off-target risks and side effects. He also highlights that DNA diffusion goes beyond simply producing DNA sequences; instead, it allows researchers to also tune key parameters like specificity, activity, and intensity.
DNA-Diffusion as an Innovation for Gene Therapy
Designing CREs systematically has always been a challenge due to their complexity. Their activity depends on transcription factor binding patterns, chromatin accessibility, and cell type. DNA-Diffusion is an AI model based on diffusion models similar to those used in image generation.
In image generation, models learn patterns at the pixel level. The team applied the same principle to DNA. Here, instead of pixels, DNA-diffusion learns from chromatin accessibility data of DNase hypersensitive sites across different cell lines. This data tells us which regions of DNA are ‘open’ and available for transcription. It generates synthetic CREs about 200 base pairs long as output, mimicking the natural transcription factor binding sites.
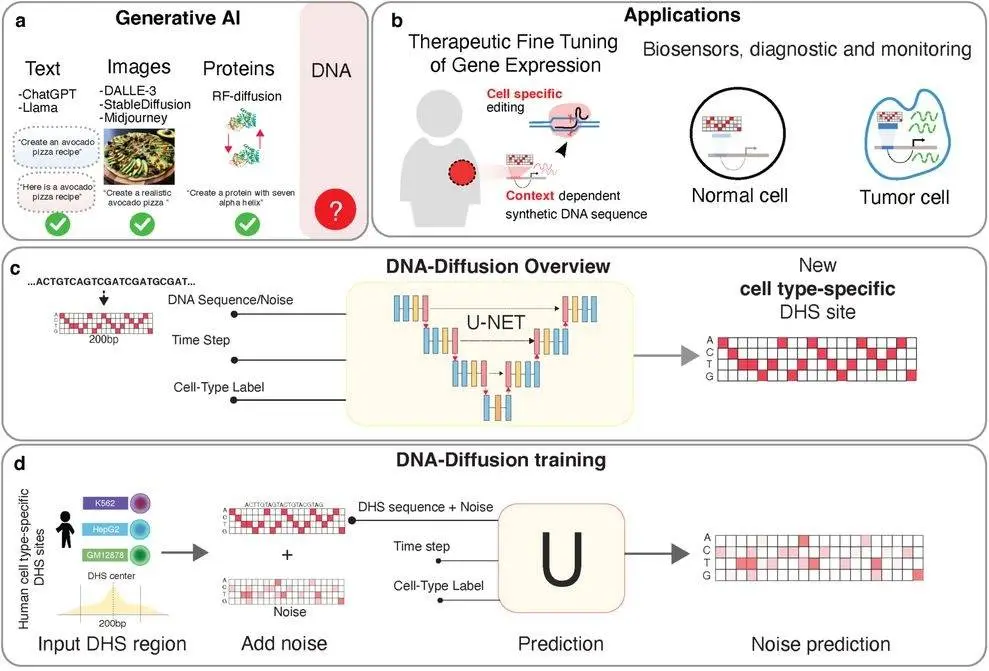
Image Source: https://doi.org/10.1101/2024.02.01.578352
Therapeutic Promise of DNA-Diffusion: Targeting a Cancer Gene
For the STARR-seq (Self-Transcribing Active Regulatory Region Sequencing), the team produced a massive library of 5850 synthetic CREs generated by the model after training. These were tested experimentally in three different cell lines. The designs showed predictable and desired activity in cells, confirming AI could generate functional regulatory DNA.
EXTRA-seq assay was used to test whether synthetic CREs can modulate genes in their natural genomic contexts. Here, the team successfully reactivated AXIN2 in its native chromosomal environment, a gene known to be protective against chronic lymphocytic leukemia (a blood cancer affecting B cells), as a validation test. This reactivation was only seen in targeted B cells and not in other cell types, demonstrating precise specificity by synthetic CREs.
This proves that generated elements can be integrated into the genome and drive real therapeutic gene expression. In fact, many of the sequences were actually more effective at turning on the AXIN2 gene than the natural regulatory elements.
How DNA-Diffusion is Outperforming Earlier Methods
Previous computational methods often produce sequences that were either strong but non-specific or specific but weak. DNA-Diffusion achieves both strong functional activity and specificity.
Also, instead of converging on repetitive motifs, it generates a set of sequences, reducing redundancy. The AI framework learns the combinatorial logic of transcription factor binding, so its designs are more reliable across different cell types.
Future Directions and Scope of the Research
Pinello’s team is broadening the model’s applications for more cell types and diseases. The goal is to use DNA diffusion as a precision control for gene expression across biological contexts.
Pinello mentioned that, by pairing genome editing tools like CRISPR and Cas9 with DNA diffusion, genes can not only be edited but also be tuned for specific tasks.
Furthermore, Adeno-associated viruses (AAVs) used to deliver therapeutics to specific cells can also be used for CREs. Once delivered, the therapeutic gene is activated only in the intended cell or tissue using DNA-Diffusion, reducing the risks of side effects further.
Article Source: Reference Paper | Abstract | bioRxiv | Code Availability: GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.
















