Predicting how tightly a drug molecule (ligand) binds to its protein target is a cornerstone of modern drug discovery. Yet, this task remains a formidable challenge due to the complexity of 3D molecular structures and the limited availability of high-quality data. In a recent study, Jakub Poziemski and Pawel Siedlecki from the Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland, explored a cutting-edge approach: harnessing Vision Transformers (ViTs) to predict protein-ligand binding affinity from 3D structural data.
Why Protein-Ligand Affinity Matters
The interactions between proteins and ligands possess a certain detail that assists in fulfilling biological functions and enables treatment therapies. Interventions, such as drug development, require the least amount of experimental procedures. Calculating binding affinity with accuracy helps optimize the processes of drug development and laboratory examinations.
For example, other scientists relied on GNNs and CNNs to solve the binding affinity calculation problem with an AI approach. CNN employs homogenous 3D grids, achieving higher accuracy in local pattern identification. GNN approaches compounds as graphs consisting of nodes and edges. While these different methods offer novel ideas, they also come with their own shortcomings. For instance, CNN can frequently overlook significant external relations, increasing the difficulty of interpreting spatial models. On the other hand, GNN suffers when trying to capture all of the three-dimensional information about the shapes of the molecules.
Enter Vision Transformers
Initially engineered for human language comprehension, transformers have matured into a plethora of domains due to their long-range attention capabilities. Vision Transformers make this architecture image-specific by patching images into smaller pieces, treating them as sequences. Thus, 3D molecular grids are a perfect fit.
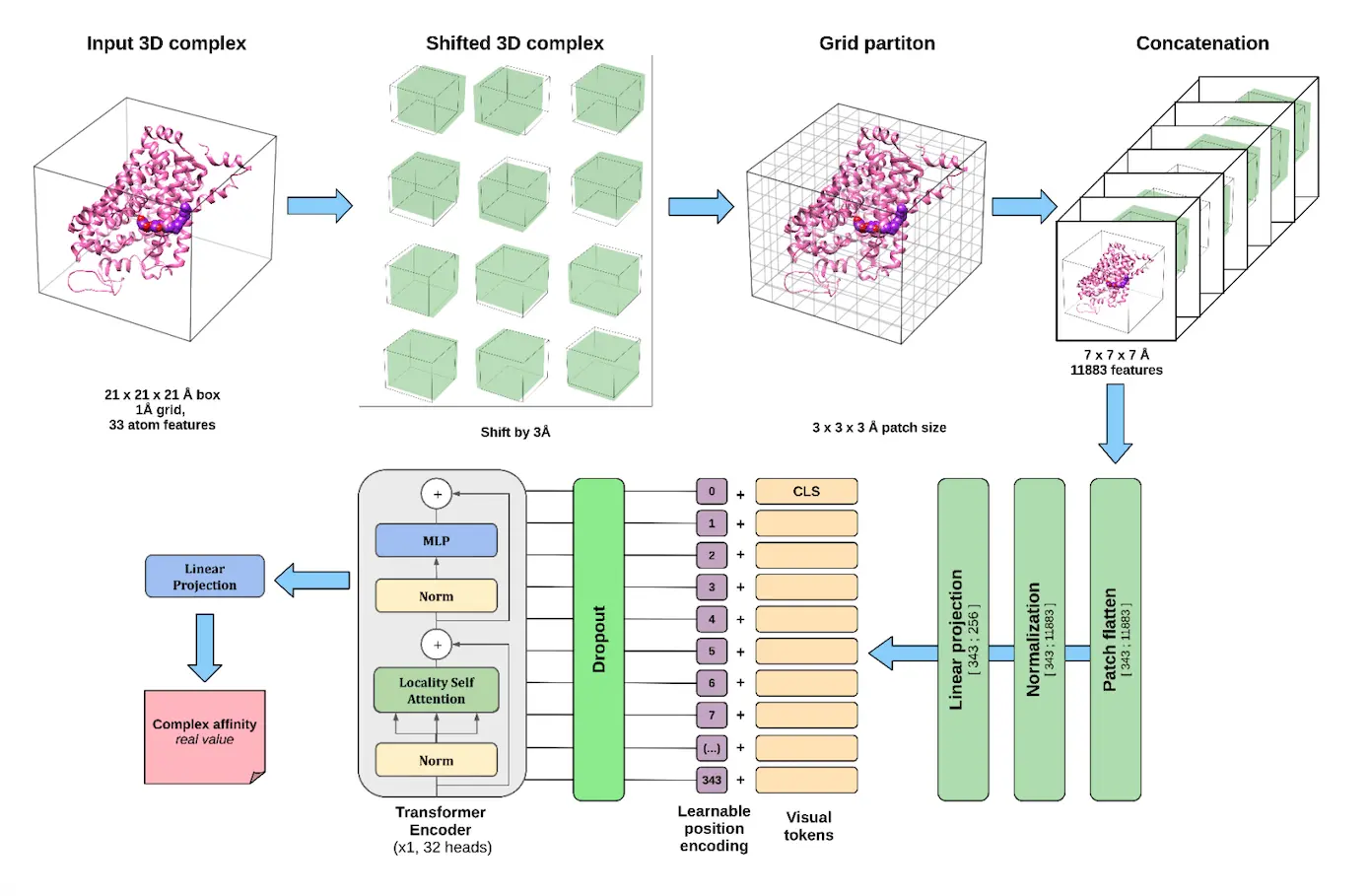
Poziemski and Siedlecki’s work takes dienes and transforms them into 3D grids containing polyhedral faces centered around ligands, with each grid acting as a three-dimensional representation of a protein-ligand complex. Each voxel contains complex features like atom types, protein secondary structure, amino acid types, pharmacophore descriptors, and others. These grids are then split into patches, which are converted to embedding vectors and processed by a transformer.
How Well Do Vision Transformers Perform?
The researchers assessed the performance of their ViT model against several competitive strategies and treated CASF2016 and CoreSet2013 datasets as benchmarks. Importantly, the ViT surpassed other grid-based models like Pafnucy and DeepDTAF, achieving top-three overall performance. With the more challenging CoreSet_2013 dataset, ViT achieved the lowest root mean squared error (RMSE) out of all the methods tested, indicating strong generalization ability.
A main takeaway from the investigation is that learnable positional encodings—those that include a learnable spatial position attached to tokens within a sequence—improve performance beyond set markings or no markings at all. This underscores the role that spatial attention plays in phenomena at the molecular level.
The Power of Data Augmentation
One of the difficulties associated with 3D molecular data is its sparsity: most grid locations are null values. To reinforce the model’s ability to learn robust patterns, the team used rotational data augmentation and produced several versions of each complex. This modification was met with considerable gains in predictability and steadily increased performance alongside the additional augmented samples.
Through ablation studies—systematically removing groups of input features—the researchers found that pharmacophore descriptors (which capture chemical features relevant to binding) were particularly important for accurate predictions. Quite remarkably, the model was able to sustain a reasonable level of accuracy even when high-level protein traits were included, indicating that features at the atomic level were extremely important.
Any prediction model loses some value without accuracy against real-world variation. The research team examines their ViT model on structure protein-ligand pairs obtained from molecular dynamics simulations, which tend to imitate the true flexibility of proteins. All deep learning models showed some degree of susceptibility towards changes in conformation, but the ViT model yielded performance relative to the other leading models.
To peek inside the “black box,” the authors used explainable AI techniques. They found that the ViT naturally focused its attention on the ligand and its immediate surroundings—the very regions most relevant for binding. Attention Score reveals clustering around the binding area, which indicates that the model is learning the right pattern and not false, chemically meaningless patterns that render no results.
Conclusion
Researchers’ study demonstrates that Vision Transformers are not just a novelty—they are powerful tools for learning complex spatial relationships in molecular data. While challenges remain, such as improving robustness to structural variability and exploring more sophisticated data augmentation, ViTs have opened new avenues for structure-based drug discovery.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.