RNAs are crucial molecules that convey genetic information necessary for life, and they have a big impact on biotechnology and medicine development. However, the large body of literature frequently slows down RNA research. To overcome this, scientists from the University of California and Georgia Institute of Technology present RNA-GPT, a multi-modal RNA chat model that uses a wealth of RNA literature to streamline RNA discovery. A strong tool for accurate representation alignment of RNA sequences, RNA-GPT makes it possible to handle user-uploaded RNA sequences quickly. Large datasets are handled, and instruction-tuning samples are produced by its scalable training workflow, which is driven by RNA-QA and automatically collects RNA annotations from RNACentral. This approach simplifies RNA research by handling complex RNA queries with ease. It also presents RNA-QA, a 407,616 RNA dataset for modality alignment and instruction tailoring.
Introduction
Powerful models may now be adapted to speed up research and drastically reduce the need for conventional experiments thanks to recent developments in the biological and medical domains. On a wide range of tasks, from long-term task planning for robotic systems to Olympiad-level mathematics and scientific reasoning, large language models (LLMs) trained on internet-scale corpora have demonstrated remarkable performance. The enormous amount of sequenced data is easily accessible, and proteins, RNAs, and DNAs may all be represented as character strings. As a result of the effective encoding of protein sequence and structure information by protein language, models such as ESM, ProteinGPT, and ProtSt were inspired.
The alignment of protein sequences and structures with textual descriptions has advanced significantly thanks to models like ProteinGPT, ProtST, ProteinChat, and ProteinCLIP, which have drawn interest from protein research and computational RNA research. However, the developments in the DNA and RNA domains are less sophisticated. The majority of prior attempts, such as RNA-FM and RiNALMo, have concentrated on particular objectives like structure and function research and promoter or enhancer prediction. One of the few models that bridge the gap between normal language and RNA comprehension, ChatNT, is more concerned with carrying out biological duties as a conversational agent than it is with offering profound RNA knowledge. It is particularly difficult to integrate several modalities, including textual descriptions, RNA sequences, and structural data when applying multimodal LLMs to RNA modeling.
Understanding RNA-GPT
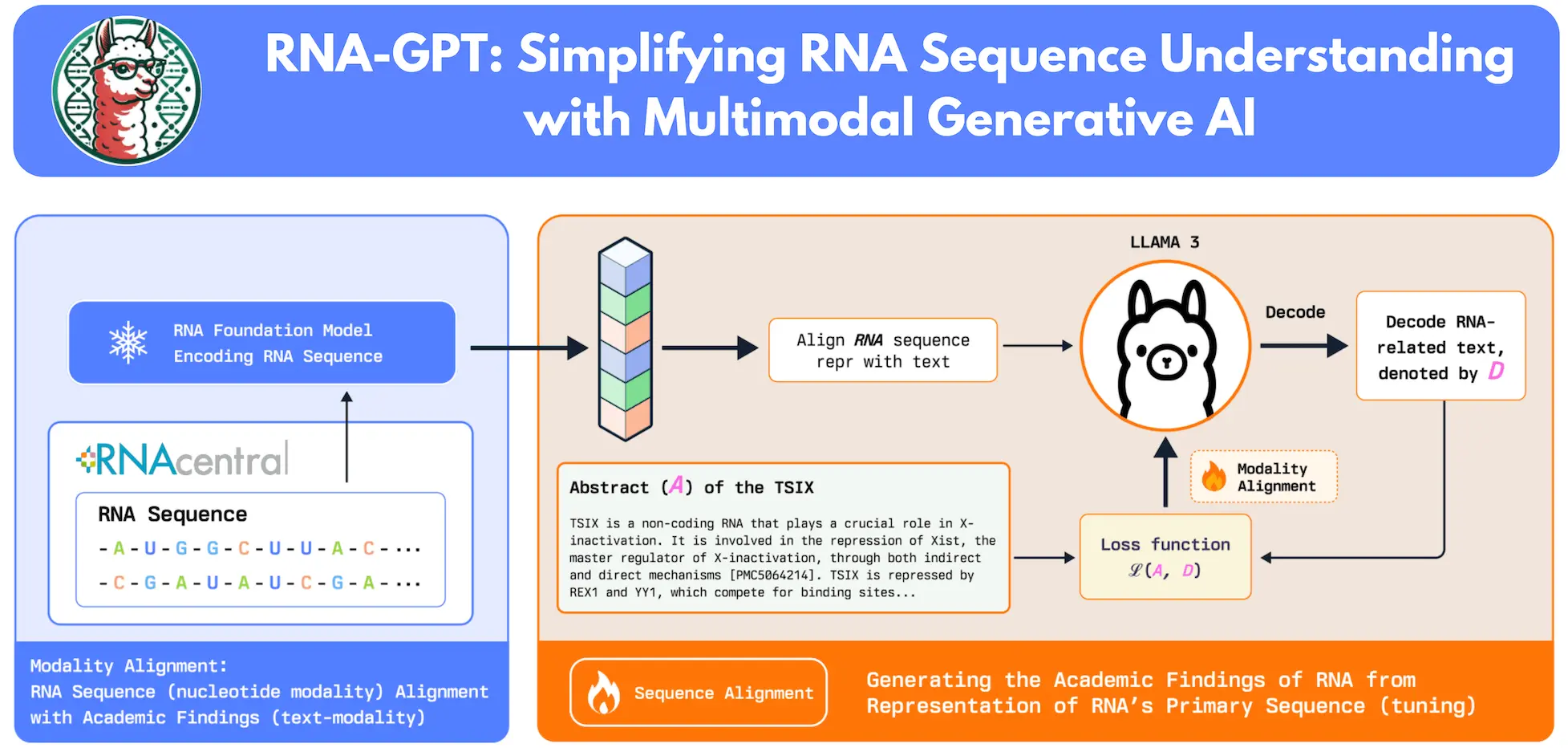
Researchers suggest a two-step method for RNA-GPT in order to get around these obstacles. First, researchers embed RNA sequences using the RNA-FM sequence encoder. Then, they use a sizable, automatically curated QA dataset from RNA Central to align these sequence representations with plain English. Second, researchers deconstruct RNA-QA’s abstract summaries into separate QA pairs for instruction tuning, which improves the model’s capacity to provide precise and understandable responses. This ensures that the model produces succinct and accurate responses. For strong general language comprehension, researchers use Meta AI’s flagship Llama-3 8B Instruction as the foundational LLM.
The study introduces RNA-GPT, a multi-modal RNA sequence conversation model that contributes to a better understanding of RNAs for biological research. RNA-QA is a collection pipeline for modality alignment instruction adjustment of RNA chat models that uses a big dataset that was taken from the RNA Central Dataset. To generate QA pairings for more than 407,616 RNA samples, the system automatically scrapes and summarizes pertinent RNA literature. For instruction tweaking, RNA-QA is a great resource because of the richness and variety of these annotations.
RNA-GPT: Methodology
Using “Lit Scan,” the study filters RNA sequences from RNA Central, yielding 420,000 RNAs with related scientific publications. To facilitate effective summarization, the sequence encoder has been improved to accommodate sequences up to 1024 nucleotides. Relevant literature is used to extract the remaining 407,616 RNAs. To reduce information loss, LDA topic modeling is used to arrange publications by topic and summarize each group separately. The token restrictions of GPT models are overcome by this divide-and-conquer strategy, which increases accuracy and manages big datasets effectively. In the final phase of summarization, these summaries are combined to construct the final annotation, which enables comprehensive, information-dense annotations of extensive RNA research profiles.
Conclusion
To improve LLM-based question-answering and speed up RNA discovery, researchers have created RNA-GPT, a multimodal conversation model for RNA sequences that offers succinct, precise answers to challenging questions. A learnable projection layer is used in the model to match RNA embeddings with natural language in LLMs such as Llama-3. Additionally, the researchers presented RNA-QA, a 407,616 RNA question-answering dataset that was compiled from a large body of RNA research literature. The scalable approach efficiently curates RNA-to-language datasets through divide-and-conquer summarization and topic modeling. Responses to experiments using the original model are encouraging, and the full RNA-QA offers even greater benefits for the creation of multimodal RNA LLMs.
Article Source: Reference Paper
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.