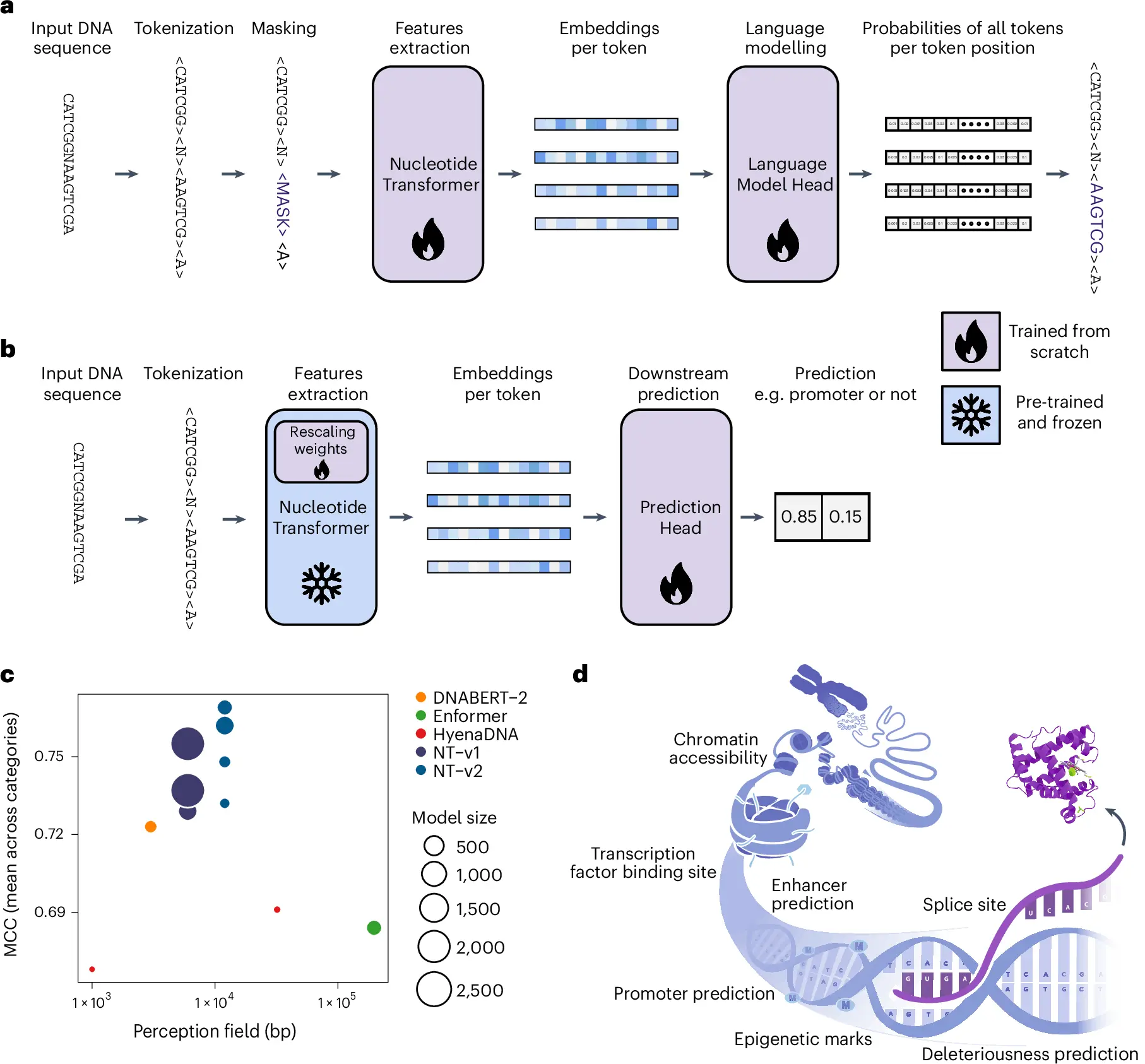
The field of genomics is ever-changing as a result of an increase in automation and aspects of biological engineering. A groundbreaking study led by researchers from InstaDeep and Nvidia introduces a Nucleotide Transformer pre-trained on DNA sequences. These models generate context-specific nucleotide representations, allowing accurate molecular phenotype predictions and improving genetic variant prioritization, marking a significant advancement in AI models.
Why Is There a Need for Foundation Models in Genomics?
Genomic information is huge, along with so many details, and continues to increase in numbers and complexity. From gene regulation to disease variants, researchers have a lot of data to sift through, process, and uncover meaning out of it, and many challenges on how such data should be interpreted are posed. A large number of complex datasets are sometimes out of the abilities of conventional machine learning techniques. Other approaches to solving such intractable problems are foundation models that build on transformers designed originally for language processing.
The Nucleotide Transformer: An Overview
The Nucleotide Transformer (NT) model was developed to process nucleic sequences efficiently and accurately (DNA/RNA). It is based on the transformer architecture, which has been successfully utilized in various fields, including language, image processing, and protein structure prediction. The researchers sought to capture useful patterns and relationships in nucleotide sequences by applying such architecture to genomic data.
Key Objectives of the Study
- Enhancer Activity Prediction: One of the major aims of the research work was to predict the enhancer activities, which are important for epigenetics. Enhancers contribute to developmental and gene expression, and their accurate prediction helps in understanding the regulatory mechanisms.
- Genomic Element Analysis: The NT model investigated various genomic components, including exons, introns, promoters, enhancers, or transcription factor binding sites. Knowledge of how these elements interact is important for basic biology and disease-related research.
- Functional Variant Prioritization: The rims functional variant prioritization was examined whereby SNPs (single nucleotide polymorphisms) were focused on their ability to regulate genes, cause disease, and the usefulness of the genome.
Methodology: Building and Training the Nucleotide Transformer
The NT model was constructed with the assistance of the dataset containing more than 484000 DNA sequences focusing on their quantitative enhancer activity towards developmental and housekeeping promoters, where each sequence comprised 249 nucleotides in total. Each of these models had two regression heads, which further assisted in the simultaneous prediction of enhancer activities, which was pertinent in optimally training the model.
The methodology followed a multilabel classification approach where each enhancer activity value was discretized into 50 evenly spaced values. By modeling enhancer activities as a regression, the model was able to narrow its prediction in terms of numerous activities and related performance metrics.
Performance Evaluation
Several methods were employed to evaluate the NT model:
- t-SNE Projections: To achieve the 2D visualization, the model embeddings were firstly subjected to t-distributed stochastic neighbor embedding, after which it was analyzed. This enabled researchers to identify different genomic elements based on how greatly they were distinguished apart from each other within the embedding space.
- Reconstruction Accuracy and Perplexity: The NT model was able to reconstruct masked nucleotide sequences. To determine the accuracy of the predictions made by the model, either the central token or random positions in the sequences were masked, and the inclusion of these tokens in the predictions was computed. Lower perplexity scores indicated better performance, signifying the model’s strong ability to reconstruct sequences.
- Attention Maps: Another interesting point was the distribution of attention across different genomic elements. With the NT model, there was an improvement in the ability to attend to relevant regions like promoters and enhancers by using heads to track these regions.
Functional Insights from Variant Prioritization
In addition to predicting enhancer activities, the NT model was fine-tuned to assess the functional impacts of genetic variants. Through single nucleotide polymorphisms (SNPs) and their functions in gene regulation, the researchers classified variants into positive and negative sets based on their effects. The zero-shot scores – including L1 distance, cosine similarity, and dot-product, among many other descriptive measurement concepts – gave quantitative insight into how changes brought by variants could impact specific genomic features.
Additionally, the NT’s performance was also assessed relative to methods such as CADD, GERP, and DeepSEA. Combining predictions from NT and these external tools provided an opportunity for the researchers to confirm that there were functional variants predicted by the model correctly.
Data Accessibility and Open-Source Contributions
One of the highlights of this research is its open-source approach. The NT model, pre-training datasets, and downstream benchmarks have been made available on platforms like HuggingFace and WashU Epigenome Browser. Such openness guarantees that the model can be adapted for more genomic research trees, as well as that the previous findings can be further exploited by the scientific community.
Conclusion
The Nucleotide Transformer has redefined the expectations of genomic data analysis. The proposed model is expected to transform the research in genomics by enabling the understanding of the biological details, the prediction of enhancer activities, and the selection of useful variants. Considering the strong methodology and ease of accessibility, this foundation model will most likely be a game changer in the arena of genomic research and clinical genomics.
Article Source: Reference Paper | The pre-trained transformer model code, weights, and inference code in Jax are available for research on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.