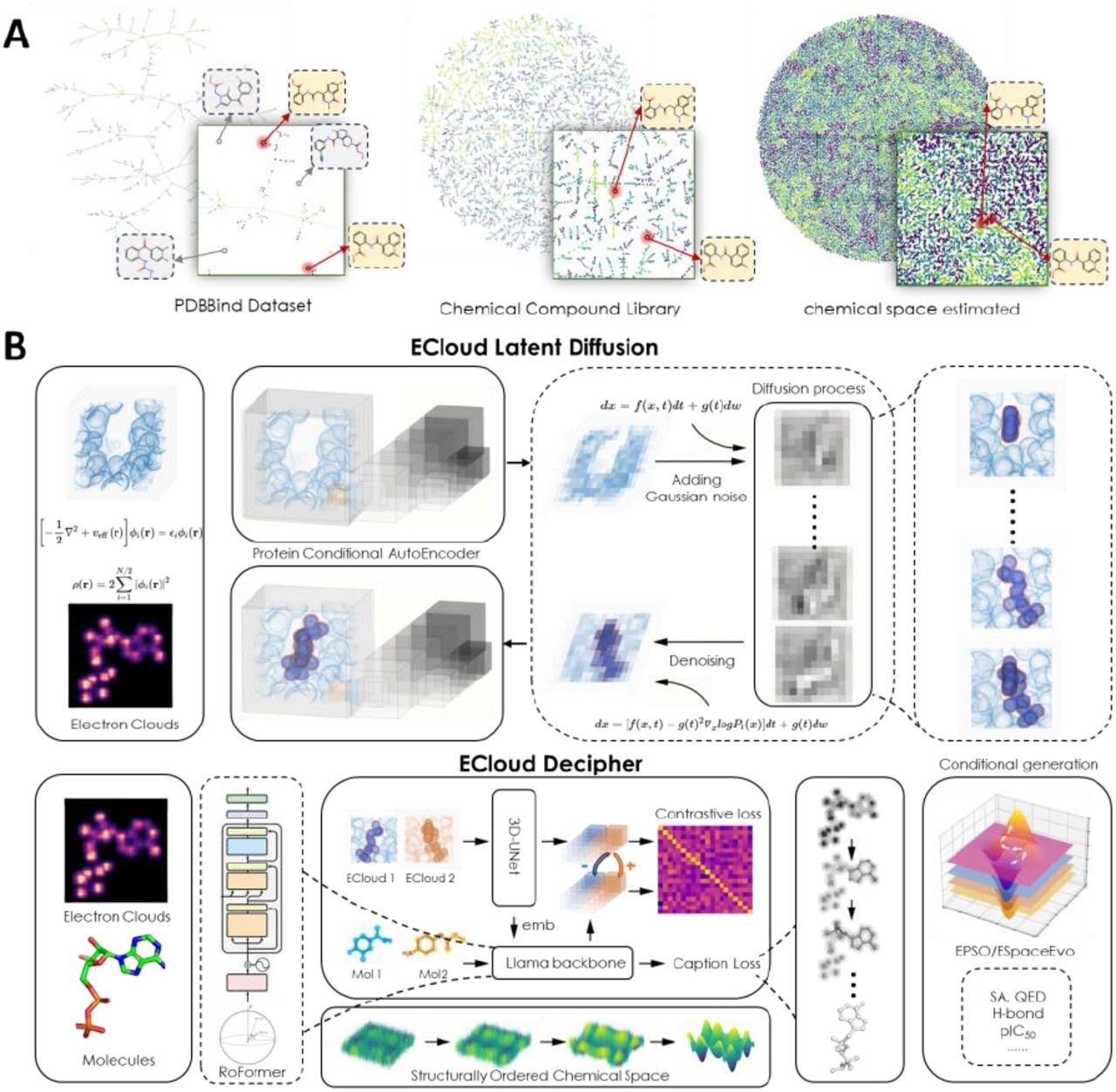
In a groundbreaking step toward more effective molecular design, researchers from Zhejiang University and others have introduced ECloudGen, a cutting-edge framework that integrates quantum chemistry with deep learning. This innovative approach focuses on utilizing electron clouds as latent variables to unlock new possibilities in structure-based molecular design.
Understanding the Role of Electron Clouds in Chemistry
Quantum chemistry deals with how electrons are positioned and arranged around the nucleus of an atom and relies on the concept of electron clouds. With quantum physics, core models of molecules as spheres and sticks are less important, as electron clouds create an abundance of worth. This explains why, for instance, they are widely used for molecular property prediction, drug design, and exploration of chemical space, among others.
RoFormer: The Backbone of ECloudGen
ECloudGen constructs itself on the RoFormer architecture, which is a modified version of the transformer that uses Rotational Position Encoding (RoPE). This backbone, based on the Llama framework, allows the system to model complex 3D geometry of electron clouds and the geometry of chemicals. The RoFormer layer solves this issue by utilizing rotational attention and analyzing the electron clouds, thus converting quantum data into molecular information.
Generating Molecules with Protein Context
An interesting and distinctive feature of ECloudGen is its capability to create molecules based on both electron cloud data and protein context. This is done through a decoder model, RoFormerd, that incorporates protein-guided cross-attention. In the absence of protein data, a protein vector can step in. Molecular captions, which are basically sequences representing a molecule, are afflicted during training to minimize the prediction error in the captioning process, i.e., prediction risk is reduced.
CEMP: Organizing Chemical Space
To assist the system in coping with the wide variety of chemical structures, the researchers resorted to a novel approach called CEMP. This method arranges the chemical space by grouping similar molecules and putting away those that are not. CMC leverages contrastive learning techniques such as CLIP; therefore, CEMP creates an organized structure where molecular spatial distribution reflects molecular structural similarities. This results in a chemical space that fosters not only the generation of molecules but also the high-fidelity optimization of the molecules.
Molecular Optimization with EPSO
The molecular optimization of a compound is arguably one of the most important tasks during the drug design phase. To avoid such an inconvenience, ECloudGen integrates a particle swarm optimization (PSO) module referred to as ECloudGen-PSO (EPSO). The optimization doesn’t depend on the models and works in the chemical space that has been learned. Hence, it is model-independent. This enables them to fine-tune the desired characteristics of the molecule without requiring a comprehensive degree of granular insight from other achieved models. The evolution of molecules through a variety of small steps provides the desired outcome, such as stronger binding affinity and a better pharmacokinetics profile.
Electron Clouds: The Core of ECloudGen
The clouds of electron density are one of the frameworks of ECloudGen, and they are used to provide an overall perspective on inter-atomic and molecular forces. The GFN2-xTB method is used to generate these clouds, which is a semi-empirical quantum mechanical method that is both cost-effective and accurate. In this way, ECloudGen can combine theoretical quantum chemistry calculations with the design of practical molecules.
Evaluating Molecular Properties
Additionally, quantifications of molecular qualities are included in the model. For instance, it quantifies the drug-like properties using the Quantitative Estimate of Drug-likeness (QED), focuses on synthetic accessibility with the help of the Synthetic Accessibility (SA) score, and examines hydrophobicity with the help of a LogP value. Besides, ECloudGen uses Lipinski’s Rule of Five to analyze whether the drug candidates selected can be successful during clinical trials.
Future Prospects of ECloudGen
Along with its technical aspects, ECloudGen also expands the horizons of drug development and materials science. The framework has the ability to enhance the drug development process by screening large volumes of chemical compounds to create precision-designed new drugs. Its modeling of electron clouds as latent variables allows each molecular phenomenon to be analyzed and predicted.
Article Source: Reference Paper | The data and source code of this study are available freely on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.