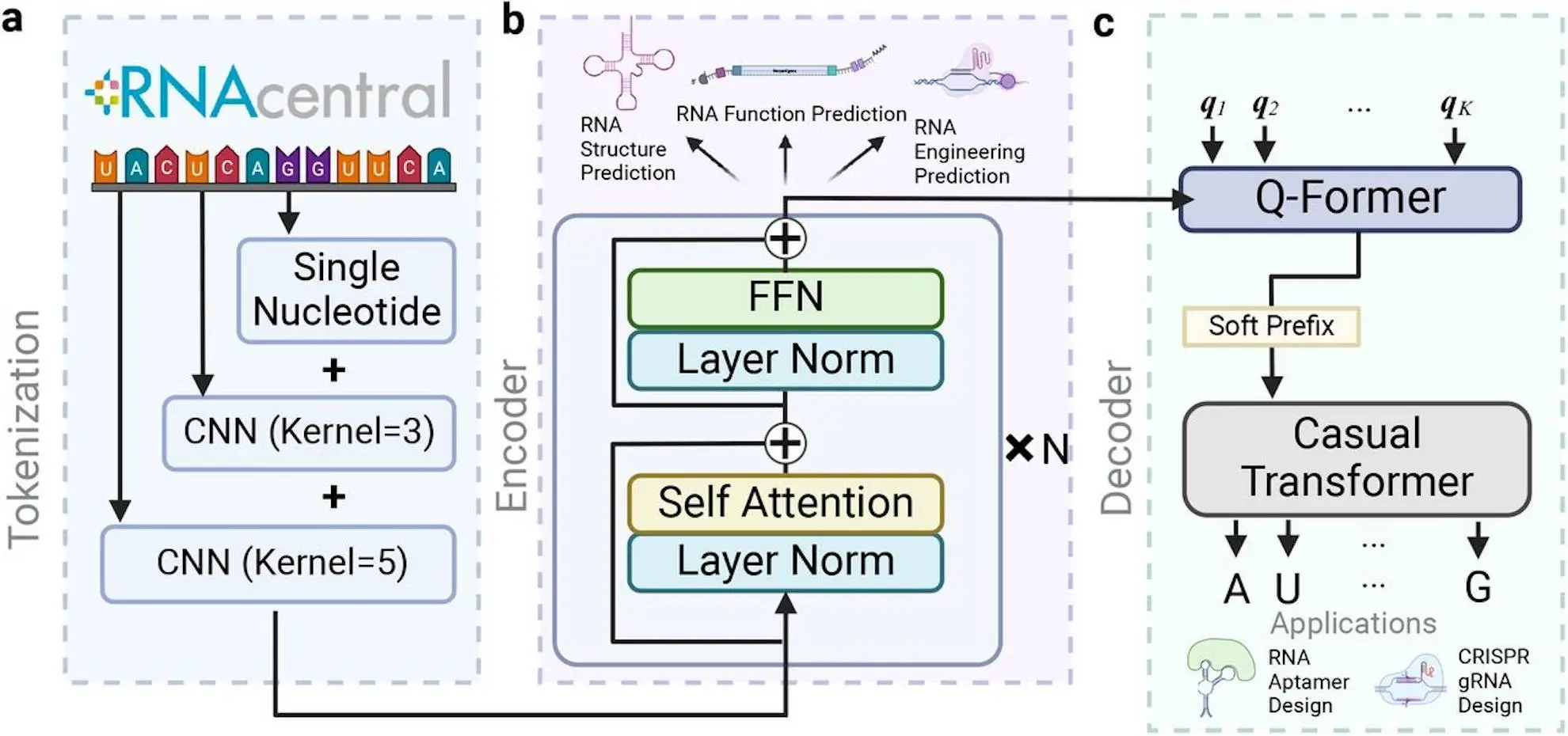
Many biological processes depend on RNA engineering, which advances the knowledge of life’s mechanisms and propels the discovery of new drugs. New strategies are made possible by improvements in RNA foundation models, but current techniques are still unable to produce unique sequences with particular functionalities. Here, scientists from Princeton University and BioMap Research present RNAGenesis, a foundation model that integrates de novo design via latent diffusion with an understanding of RNA sequences. With its Bert-like Transformer encoder, Query Transformer, and autoregressive decoder, RNAGenesis is a potent tool for deciphering RNA sequences effectively. It uses learned representations to reconstruct RNA sequences, and it performs best in nine of the thirteen benchmarks, especially when it comes to RNA structure prediction. RNAGenesis is an important tool for RNA-based therapies and biotechnology since it is also very good at creating natural-looking aptamers and tailored CRISPR sgRNAs.
Introduction
It is essential to comprehend the critical role that RNA plays in biological processes, such as protein synthesis and enzyme activity, to comprehend the intricacies of cellular functions and illnesses. Non-coding RNAs (ncRNAs) are important in these processes, supporting vital cellular processes such as signal transduction, metabolism, and transport. Since RNA can be synthesized and programmed, it is a desirable option for improvements in drug design and therapeutic development. The adaptability of RNA molecules in gene editing and therapeutic applications is further demonstrated by using aptamers and CRISPR single guide RNA (sgRNA). As a result, creating thorough RNA foundation models is crucial to furthering both clinical and scientific applications.
For extensive downstream tasks, the RNA Foundation has created several models pre-trained on large amounts of RNA sequence data to gain evolutionary information. Masked Language Modeling (MLM) techniques and encoder-only transformer frameworks, including RNA-FM and RiNALMo, are used by these models. By tokenizing RNA types for type-guided fine-tuning and using RNA motifs as biological priors, RNAErnie improves pretraining by pushing the boundaries of model size (1.6B) and establishing new standards on an extensive variety of RNA jobs, AIDO.RNA scales these developments. UTR-LM and CodonBERT are two examples of customized RNA Foundation models that concentrate on particular RNA types to identify important regulatory motifs and comprehend codon usage bias and how it affects translation efficiency.
Understanding RNAGenesis
The proposal in this research is RNAGenesis, which combines latent diffusion with de novo sequence design and RNA sequence comprehension. To effectively capture context information with several granularities, the encoder uses hybrid N-gram tokenization, which is a Bert-like Transformer. A set of fixed-length latent vectors is then created by compressing the Encoder’s representations using a Query Transformer. To capture the distribution of RNAs in latent space, researchers train a score-based denoising diffusion model, and the autoregressive decoder can reconstruct RNA sequences from these latent variables. In addition to performing exceptionally well on RNA sequence comprehension tests (best in 9 out of 13), extensive experiments demonstrate that RNAGenesis is highly skilled at creating CRISPR single guide RNA (sgRNA) and natural-looking RNA aptamers.
Limitation and Future Work
Although RNAGenesis makes significant strides in modeling and creating RNA sequences, several intrinsic drawbacks should be noted. First, the model only works with individual RNA sequences at this time, which might not fully take advantage of the larger context that RNA structure or functional annotations offer.
Multi-modal models that combine sequence data with structural information (like secondary and tertiary structures) and functional annotations (like gene expression or RNA modifications) may be investigated in future research. A more thorough understanding of RNA function and its role in cellular processes may be made possible by such multi-modal frameworks.
The creation of a single model that can manage both coding and non-coding RNAs at the same time is another possible direction for advancement. Though separate models for coding and non-coding RNAs could be needed to capture the special traits and roles of various kinds of RNAs, RNAGenesis is currently tuned for generic RNA sequencing tasks. Across multiple RNA types, a single model that can distinguish between coding and non-coding sequences could increase prediction accuracy and expedite processes.
Lastly, expanding RNAGenesis to a unified model that can manage RNA, DNA, and protein sequences in a single framework would be a fascinating future direction. Researchers may examine the complicated relationships between these basic biological molecules using such a unified model, which would also provide light on delicate processes like the core dogma, protein-RNA interactions, and the regulation of gene expression.
Conclusion
An important breakthrough in RNA sequence generation has been made with the creation of RNAGenesis, which provides a strong framework that combines de novo design skills with an understanding of RNA sequences. The model addresses significant shortcomings of current techniques by utilizing latent diffusion, making it possible to construct highly selective functional RNA molecules. Many benchmarks show that it performs better in both sequence design and understanding, especially in the production of CRISPR sgRNAs and aptamers. These findings highlight how RNAGenesis has the potential to propel advancements in synthetic biology, biotechnology, and RNA-based therapies while offering a useful instrument for expanding RNA engineering across a range of applications.
Article Source: Reference Paper | The source code to replicate the results of this study is available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.