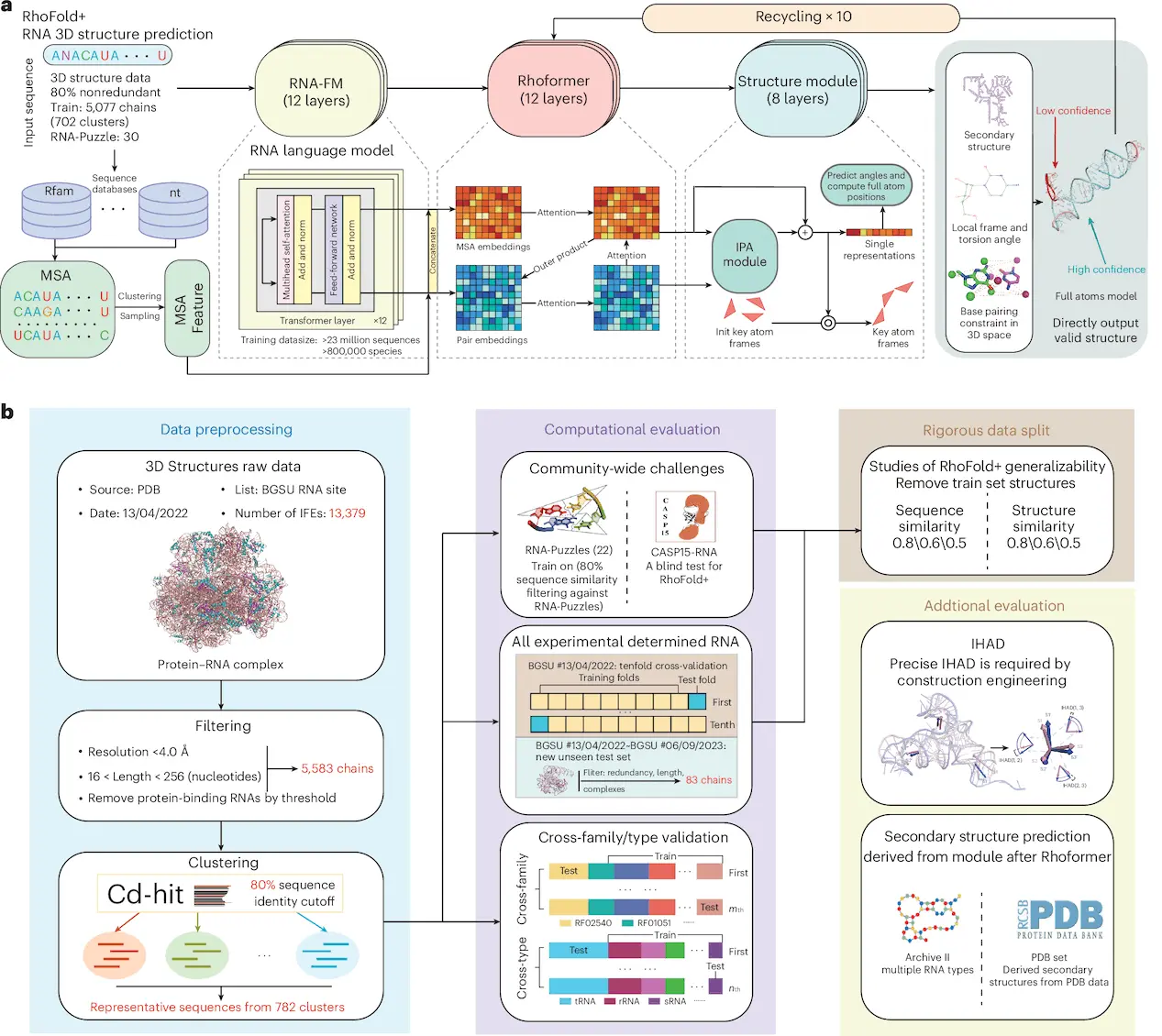
Although RNA 3D structures are important in understanding their functions and in the design of new drugs, their accurate prediction is still a challenge. Structural flexibility makes computational predictive efforts even more problematic because of the scarcity of experimental input. In this study, researchers from The Chinese University of Hong Kong, Harvard University, Fudan University, and Shanghai Zelixir Biotech Company Ltd. presented RhoFold+, a deep learning method based on RNA language modeling to faithfully predict the 3D structures of single-chain RNAs from samples. The RNA language model, complemented with strategies to deal with data scarcity, makes RhoFold+ a fully automated pipeline for RNA 3D structure prediction. It beats state-of-the-art techniques, including human expert groups, for RNA-Puzzles and CASP15 natural RNA targets. The properties of RhoFold+ to predict RNA secondary structures and interhelical angles are testable, thus extending its application to studies of RNA structure and function.
Introduction
The fundamental tenets of molecular biology rest on RNA molecules. The effects of the geometry of RNA on the regulation and function of the gene have been well studied. It was also suggested that RNA targeting is likely to emerge as a powerful synthetic biology design element and an important target druggable for drug development. Of the entire human genome, only 3% codes for proteins, whereas more than 85% is transcribed; yet, this states how many transcribed RNAs are there without defined structure and function. Often, high-resolution structural details can enable a better predictive understanding of the relevant RNA molecules.
The great flexibility of conformational states that RNA molecules possess makes it experimentally difficult to determine 3D structures of RNA. Till December 2023, RNA-containing complexes were 2.1% of the nearly 214,000 PDB structures, as opposed to less than 1.0% for only RNA structures. Even with advancements in NMR spectroscopy, cryogenic electron microscopy, and X-ray crystallography, these methods’ low throughput is limited and is directed toward specific requirements. Computational techniques to be aided with RNA sequence data have developed into a complementary approach for predicting the 3D structure of RNA.
These methods can be broadly classified into two major categories. The first is de novo techniques such as FARFAR2, 3dRNA, and SimRNA, which tend to be more predictive but require far-reaching large-scale sampling due to their computational demands. The second is template-based modeling methods such as ModeRNA and RNAbuilder, which are limited by the small number of templates in the library.
Looking into RhoFold+
Here, the authors present RhoFold+, a deep learning, language model-based tool for quick and accurate de novo RNA 3D structure prediction. RhoFold+ is a fully automated, entirely different version from RhoFold, which uses improved MSA integration and other features to enhance performance. The aim is to resolve single-chain structures of RNAs that are loosely interacting with any other molecules. The knowledge so acquired will enhance the understanding of RNA biology and lay the foundation for resolving more intricate questions.
Performance of RhoFold+
- RhoFold+ works quickly and well since it doesn’t require lengthy, computationally intensive sampling procedures.
- It does not even depend on expert knowledge, a crucial ingredient to the best-performing methods for RNA structure predictions so far.
- The remarkable robustness of RhoFold+ in cross-fold validation shows how it can generalize from different training datasets and accurately predict already-known as well as new RNA 3D structures.
- In addition, RhoFold+ can reliably infer hidden RNA structures even under cross-family and cross-type validation conditions.
- RhoFold+ can predict RNA secondary structures well, even if it is intended to predict 3D structures.
Limitations
Like other previous deep learning techniques, RhoFold+ suffers from constraints in RNA structure prediction despite its promising performance.
First, RNA molecules can adopt various forms with their dynamic nature and interactions with other molecules. Hence, prediction is hampered due to poor understanding of their structural diversity, such as an RNA junction, which could be understood better if treated as dynamic ensembles because they can adopt many forms.
Second, it is still very difficult, due to a lack of data, especially for sequences longer than 500 nucleotides, to anticipate large and complex RNA structures, particularly with many helices or pseudoknots.
Third, because most methodologies usually fail to take into account the above-mentioned interactions, which degrade accuracy, RNA complexes containing ligands or proteins create new complications. RNA complexes can be predicted by such techniques as AlphaFold3 and RoseTTAFoldNA, although their accuracy is still low, and they outperform RhoFold+ on single-strand RNAs.
Fourth, the data used to train RhoFold+ and similar models derive from a specific set of environmental conditions that translate poorly to the varied and changing solution conditions of RNAs in vivo. These conditions include different ion concentrations- that is, potassium and magnesium ions- and ligands that are well known to be crucial in RNA stability and folding.
Conclusion
This paper is about the development of RhoFold+, a deep learning-based end-to-end language model developed to predict the 3D structure of RNA from the sequence. RhoFold+ is an entirely automated model with differentiable processing, using techniques supplementing the small training data, as well as an RNA language model pre-trained on over 23.7 million RNA sequences devoid of the false leakage of structural information. Strong performance has been established by RhoFold+ concerning nonoverlapping and nonredundant RNA-Puzzles structures, achieving a mean r.m.s.d. of less than 4 Å and also superseding any other deep learning-based RNA structure prediction method on the CASP15 natural RNA targets. The utilization of RhoFold+ for predicting IHA, which is inspired by building engineering design backed by NMR and cryogenic electron microscopy, promises that it would accelerate the pace of experiment identification of more RNA structures.
Article Source: Reference Paper | RhoFold+ is also freely available for academic purposes on this server | GitHub Link.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.