Scientists from the Technical University of Denmark (DTU) and the University of California San Diego present a systems biology workflow that integrates genome mining with a detailed pangenome analysis for detecting genes associated with a particular biosynthetic gene clusters (BGCs). The study will help find novel secondary metabolites, comprehend their physiological functions better, and find and examine gene sets related to BGC.
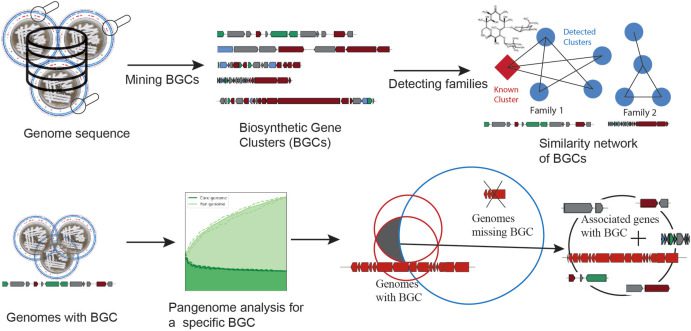
Image Source: Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets
In silico, genome mining provides easy access to secondary metabolite biosynthetic gene clusters encoding the biosynthesis of many bioactive compounds, which are the basis for many important drugs used in human medicine. However, the association between BGCs and other functions encoded in the genomes of producers has remained elusive.
In-depth computational methods that investigate bacterial DNA have been developed over the past 20 years. These models search for intriguing metabolites (molecules involved in metabolism) that cause a potent biological response. Their effects could be harmful or positive, such as influencing the creation of novel antibiotics, anti-cancer medications, or bio-based insecticides for use in agriculture.
The computer techniques look for distinct genetic markers around DNA that produce different molecules of interest in medicine, agriculture, and industry. The connection between these genomic areas and the larger bacterial systems is still unknown.
An innovative computational method for analyzing thousands of bacteria DNA sequences has been created by the research team. The findings of the research were published in KeAi journal Synthetic and Systems Biotechnology.
According to Tilmann Weber, one of the study authors and an Associate Director of the Natural Product Genome Mining group at the Novo Nordisk Foundation Center for Biosustainability at DTU (DTU Biosustain), the objective of the study was to determine which genomic parts work in unison to produce valuable compounds.
The broad bacterial family Enterobacteriaceae contains both harmful bacteria like common infectious pathogens Salmonella and E-coli and harmless bacteria that coexist in symbiosis with other living beings. Surprisingly, the computational analysis carried out for this work revealed a huge number of previously undiscovered gene clusters in charge of metabolites of potential interest. It is yet to understand how these substances interact with human hosts and other environments, as well as how they contribute to the growth of bacteria.
According to research, various gut microorganisms produce colibactin, a chemical that has been linked to colon cancer. The researchers in this study identified a number of genetic components that are constant in colibactin-containing bacteria.
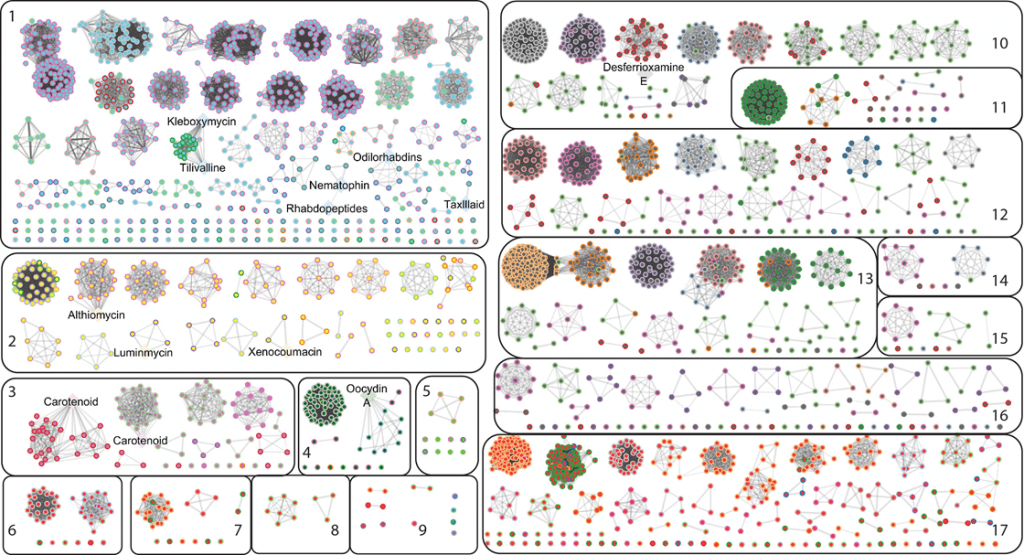
Image Source: Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets
According to the first author of the study Omkar Mohite, who is a postdoctoral researcher at DTU Biosustain, a list of biological components that work together to enable the generation of the genotoxin that can cause colon cancer may be predicted using these related signatures, which is useful information that may help to advance future treatment approaches.
The drive to understand how biological systems are made up of several parts interacting together is what got me into science. I believe that this puzzle can be solved with new approaches to the integration and investigation of large numbers of datasets, like the approach we’ve used in this study.
Omkar Mohite
The researchers showcase a systems biology methodology that combines pangenome analysis and genome mining techniques and makes use of an increasing amount of genome sequences.
The method developed can help to examine the potential for the biosynthesis of secondary metabolites in a vast number of bacterial genomes and find unique connections between secondary metabolism and other genome functions.
The current approach can be easily expanded to actinobacteria, and other microorganisms that produce a wide variety of secondary metabolites with antibiotic and other therapeutic and industrial properties as sequencing technology develops and genome availability rises.
With the aid of such an approach, we might be able to pinpoint important enzymes or regulators that are always associatively active when a particular antibiotic encoded BGC is present.
Story Sources: Mohite, O. S., Lloyd, C. J., Monk, J. M., Weber, T., & Palsson, B. O. (2022). Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets. Synthetic and Systems Biotechnology, 7(3), 900–910.
DOI: https://doi.org/10.1016/j.synbio.2022.04.011
https://www.keaipublishing.com/en/news/study-highlights-undiscovered-potential-of-bacterial-compounds-and-genes-linked-to-colon-cancer-related-toxin/
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.






